Difference between revisions of "General Information/Transposase expression and activity"
Francislon (talk | contribs) m (Text replacement - "http://tncentral.ncc.unesp.br/cgi-bin/tn_report.pl?id=" to "https://tncentral.ncc.unesp.br/report/te/") |
|||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
==Transposase expression and activity== | ==Transposase expression and activity== | ||
| − | While many of the classical mechanisms of controlling gene expression, such as the production of transcriptional repressors (IS''1'': <ref | + | While many of the classical mechanisms of controlling gene expression, such as the production of transcriptional repressors ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']: <ref><pubmed>2553980</pubmed></ref><ref name=":0"><pubmed>1848178</pubmed> |
| + | </ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=\IS2 IS''2'']: <ref><pubmed>8107136</pubmed></ref> or translational inhibitors (anti-sense RNA in the case of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']; see <ref name=":1"><pubmed>8556869</pubmed> | ||
| + | </ref> are known to operate in Tpase expression, several other mechanisms have also been uncovered. | ||
===Impinging transcription=== | ===Impinging transcription=== | ||
| − | Many ISs have evolved mechanisms which attenuate their activation by impinging transcription following insertion into active host genes. This was originally observed in the case of IS''1''<ref | + | Many ISs have evolved mechanisms which attenuate their activation by impinging transcription following insertion into active host genes. This was originally observed in the case of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']<ref name=":2"><pubmed>6281761</pubmed> |
| + | </ref><ref><pubmed>6313938</pubmed></ref>, for [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]]<ref name=":3"><pubmed>3015876</pubmed> | ||
| + | </ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']. To our knowledge other elements have not been examined. | ||
This effect may be the result of disrupting complexes between Tpase and cognate DNA ends and could reflect either inhibition of transposase binding or disruption of extant transposase-end complexes. | This effect may be the result of disrupting complexes between Tpase and cognate DNA ends and could reflect either inhibition of transposase binding or disruption of extant transposase-end complexes. | ||
| − | In the case of [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]], transcription originating from within the element and impinging on the left end has also been shown to reduce activity<ref | + | In the case of [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]], transcription originating from within the element and impinging on the left end has also been shown to reduce activity<ref name=":3" />. It is possible that transcription disrupts the formation of intermediates including transposase and one or both Mu ends which lead to stable transposition complexes. |
===Sequestration of translation initiation signals=== | ===Sequestration of translation initiation signals=== | ||
| − | One such mechanism observed with IS''10'' and IS''50'', and potentially present in several other ISs, is the sequestering of translation initiation signals in an RNA secondary structure [[:File:1.32.1.png|(Fig. | + | One such mechanism observed with [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''], and potentially present in several other ISs, is the sequestering of translation initiation signals in an RNA secondary structure [[:File:1.32.1.png|(Fig.21.1)]]. These ISs carry inverted repeat sequences located close to the left end which include the ribosome binding site or translation initiation codon for the Tpase gene. Transcripts from the resident Tpase promoter include only the distal repeat unit which is unable to form the secondary structure, while transcripts from neighboring DNA include both repeats and would generate secondary structures in the mRNA which would sequester translation initiation signals<ref><pubmed>2416461</pubmed></ref><ref name=":4"><pubmed>2438419</pubmed> |
| + | </ref>. This has been demonstrated experimentally for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''] but several additional insertion sequences carry such potential structures and might be expected to exhibit a similar mechanism (see <ref><pubmed>7934941</pubmed></ref>). | ||
| − | [[Image:1.32.1.png|thumb|center| | + | [[Image:1.32.1.png|thumb|center|680x680px|'''Fig.21.1.''' Sequestration of translation initiation signals. The figure shows the left end of an IS (green) with its terminal inverted repeat (red). A transcript impinging from outside the element (initiated at P extern) is shown as a wavy line above, and transcription driven by the indigenous promoter (PTpase) is indicated below. Internal inverted repeat sequences are indicated by bold lines, and their relative orientation is shown by arrows. The bottom panel shows the sequence and secondary structure features of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS10] in this region. |alt=]] |
===Programmed Translational Frameshifting=== | ===Programmed Translational Frameshifting=== | ||
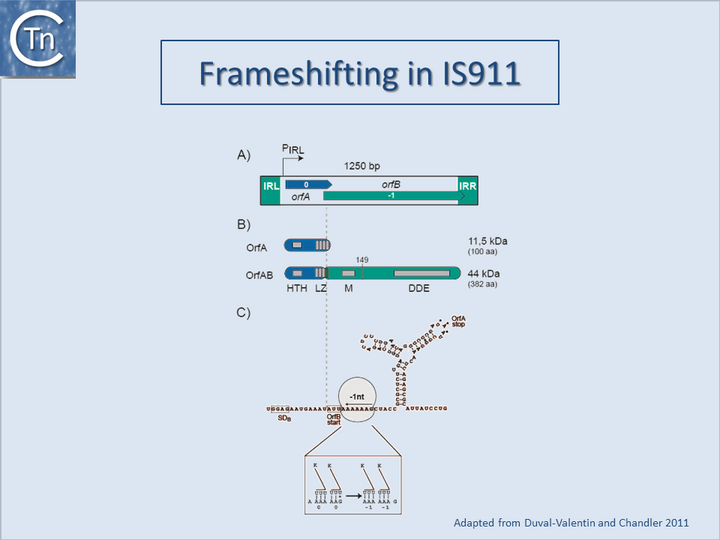
| − | A second mechanism acts at the level of translation elongation and involves programmed translational frameshifting between two consecutive open reading frames [[:File:1.32.1.png|(Fig. | + | A second mechanism acts at the level of translation elongation and involves programmed translational frameshifting between two consecutive open reading frames [[:File:1.32.1.png|(Fig.21.1)]]. Typically a -1 frameshift is observed in which the translating ribosome slides one base upstream and resumes in the alternative phase. This generally occurs at the position of so-called slippery codons in a heptanucleotide sequence of the type '''X XXZ ZZN''' in phase 0 (where the bases paired with the anticodon are shown as triplets) which is read as '''XXX ZZZ N''' in the shifted -1 phase [[:File:1.32.1.png|(Fig.21.2)]] (see e.g. <ref name=":5"><pubmed>8384687</pubmed> |
| − | <center> | + | </ref><ref><pubmed>8852897</pubmed></ref><ref><pubmed>8811194</pubmed></ref>, http://recode.genetics.utah.edu/). The sequence '''A AAA AAG''' is a common example of this type of heptanucleotide. Ribosomal shifting of this type is stimulated by structures in the mRNA which tend to impede the progression of the ribosome such as potential ribosome binding sites upstream or secondary structures (stem-loop structures and pseudoknots) downstream of the slippery codons. Translational control of transposition by frameshifting has been demonstrated both for [[IS Families/IS1 family|IS''1'']]<ref><pubmed>2543983</pubmed></ref><ref name=":0" />, and for members of the [[IS Families/IS3 family|IS''3'' family]] [[:File:1.33.2.png|(Fig.21.3)]] (<ref><pubmed>1660923</pubmed></ref>; see also <ref name=":5" />) but may also occur in several other IS elements (see for example [[IS Families/IS5 and related IS1182 families|IS''5'']] and [[IS Families/IS630 family|IS''630'' families]]). For [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] and members of the [[IS Families/IS3 family|IS''3'' family]], the upstream frame appears to carry a DNA recognition domain whereas the downstream frame encodes the catalytic site. While the product of the upstream frame alone acts as a modulator of activity, presumably by binding to the IR sequences, frameshifting assembles both domains into a single protein, the Tpase, which directs the cleavages and strand transfer necessary for mobility of the element. The frameshifting frequency is thus critical in determining overall transposition activity. Although it has yet to be explored in detail, frameshifting could be influenced by host physiology thus coupling transposition activity to the state of the host cell. |
| − | + | <center>[[Image:1.33.1.png|thumb|center|720x720px|'''Fig.21.2.''' Programmed translational frameshifting schema. '''A)''' Programmed translational frameshifting. The IS is shown in green and the terminal IRs in red. Two consecutive open reading frames (A and B) together with their relative reading phases (0 and –1, respectively) and the region of overlap are shown within the IS element in yellow. The overall secondary structure of the corresponding mRNA is shown below (bold line). The group of codons that permit the ribosome to slide back one nucleotide is also indicated by an open box. The potential to form a pseudoknot is also indicated. Both the secondary structure and pseudoknot slow down the ribosome and contribute to the frequency of translational frameshifting. The bottom of the figure shows how frameshifting can assemble two different functions into one protein. Here, this occurs between the N-terminal region that carries a helix–turn–helix motif ('''HTH''') which allows sequence-specific DNA binding to the ISs as well as multimerization motifs (not shown) and the C-terminal region including the '''DD(35)E motif''' which is essential for catalysis. '''B)''' A general schematic showing the ribosome A, P and E sites, the slippery codons and the downstream secondary structure. |alt=]]<br />[[Image:1.33.2.png|thumb|center|720x720px|'''Fig.21.3.''' Frameshifiting in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''] (''[[IS Families/IS3 family|IS3 family]]''). | |
| − | |||
| − | ''' | + | '''A)''' Organization of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911'']. Top: Left (IRL) and right (IRR) terminal inverted |
| − | |||
| − | ''' | + | repeats in green; ''orfA'' (blue) and ''orfB'' (green) reading frames; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''] promoter, pIRL. |
| − | + | '''B)''' Structure/function map of OrfA and OrfAB; HTH: helix-turn-helix motif for DNA | |
| − | |||
| − | '''B) | ||
recognition and binding; LZ: leucine zipper motif involved in OrfAB and OrfA | recognition and binding; LZ: leucine zipper motif involved in OrfAB and OrfA | ||
| Line 35: | Line 36: | ||
catalytic domain. | catalytic domain. | ||
| − | '''C) | + | '''C)''' Schematic of the stem-loop which facilitates ribosome slowdown and slippery codons at which the ribosome undergoes rephasing in the -1 direction.|alt=]] |
| − | |||
| − | |||
| − | |||
</center> | </center> | ||
===Programmed Transcriptional Frameshifting=== | ===Programmed Transcriptional Frameshifting=== | ||
| − | Another type of frameshifting may also occur in bacterial insertion sequences. This occurs at the transcriptional level and involves misreading and slippage of RNA polymerase on stretches of A residues. Although no real data is at present available, it may occur relatively frequently<ref | + | Another type of frameshifting may also occur in bacterial insertion sequences. This occurs at the transcriptional level and involves misreading and slippage of RNA polymerase on stretches of A residues. Although no real data is at present available, it may occur relatively frequently <ref><pubmed>PMC1088944</pubmed></ref><ref><pubmed>16460832</pubmed></ref><ref><pubmed>21673094</pubmed></ref>. |
| − | === | + | ===Recoding suppression of stop codons using the unusual amino acids Pyrrolysine and Selenocysteine=== |
| − | Another type of recoding which appears to occur in ''[[wikipedia:Methanosarcina|Methanosarcina]]'', is the “suppression” of the stop codon, UAG, by insertion of Pyrrolysine. This was first noted in the Methylamine methyltransferases which are important in the production of methane by archaeal methanogens | + | Another type of recoding which appears to occur in ''[[wikipedia:Methanosarcina|Methanosarcina]]'', is the “suppression” of the stop codon, UAG, by insertion of Pyrrolysine. This was first noted in the Methylamine methyltransferases which are important in the production of methane by archaeal methanogens <ref><pubmed>10762254</pubmed></ref> identified an in-frame amber codon (TAG) in the trimethylamine methyltransferase genes of both ''[[wikipedia:Methanosarcina_barkeri|M. barkeri]]'' and ''[[wikipedia:Methanosarcina_thermophila|M. thermophila]]''. However, at least in the case of ''M. barkeri'', abundant quantities of the full-length protein could be obtained and it appeared that the TAG codon was read as Lys. This later proved to be the unusual amino acid pyrrolysine. Several IS copies in these archaeal methanogens carry TAG codons which are presumably “suppressed” by decoding as pyrrolysine. |
| − | In this framework, other stop codons are known to be suppressed by decoding as selenocysteine <ref | + | In this framework, other stop codons are known to be suppressed by decoding as selenocysteine <ref><pubmed>15788401</pubmed></ref><ref><pubmed>23185002</pubmed></ref><ref><pubmed>15372017</pubmed></ref><ref><pubmed>12029131</pubmed></ref>. To our knowledge a single IS from the [[IS Families/IS3 family|IS''3'' family]], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISDvu3 IS''Dvu3''] from ''[[wikipedia:Desulfovibrio_vulgaris|Desulfovibrio vulgaris]]'', includes a [[wikipedia:Selenocysteine|selenocysteine]] inserted at a stop codon in its ''orfB'' frame ([https://www.ornl.gov/staff-profile/miriam-l-land M. Land] personal communication). Undoubtedly additional examples will be identified in the future. |
| − | The inclusion of these amino acids involves the presence of specific types of secondary structures in the mRNA [[:File:1.32.1.png|(Fig. | + | The inclusion of these amino acids involves the presence of specific types of secondary structures in the mRNA [[:File:1.32.1.png|(Fig.21.4)]]. |
| − | [[Image:1.33.3.png|thumb|center| | + | [[Image:1.33.3.png|thumb|center|680x680px|'''Fig.21.4.''' Insertion of non-standard amino acids. Different RNA structures which signal the incorporation of non-canonical amino acids. From left to right: '''selenocysteine insertion element (SECIS)'''. A specific factor, SelB, bridges selenocysteine loaded tRNA and SECIS. The result is that the selenocysteine is incorporated into the peptide at a UGA stop codon. Eukaryotes and archaea use a different type of structure; '''pyrrolysine insertion elements (PYLIS).''' This type of structure likely to signal for pyrrolysine to be inserted in the archaeal methyltransferase peptide at a UAG codon in a similar to SECIS. From Schimmel and Beebe (2004)|alt=]] |
===Translation termination=== | ===Translation termination=== | ||
| − | A third potential mechanism derives from the observation that the translation termination codon of Tpase genes of certain elements is located within their IR sequences. Although to our knowledge, no extensive analysis of the significance of this arrangement has yet been undertaken, it seems possible that it may in some manner couple translation termination, transposase binding, and transposition activity. The transposase gene of several elements does not possess a termination codon. These include IS''240C'', a member of the IS''6'' family (Y. Chen and J.Mahillon unpublished), two members of the IS''5'' family, IS''427''<ref | + | A third potential mechanism derives from the observation that the translation termination codon of Tpase genes of certain elements is located within their IR sequences. Although to our knowledge, no extensive analysis of the significance of this arrangement has yet been undertaken, it seems possible that it may in some manner couple translation termination, transposase binding, and transposition activity. The transposase gene of several elements does not possess a termination codon. These include [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS240C IS''240C''], a member of the [[IS Families/IS6 family|IS''6'' family]] (Y. Chen and [https://uclouvain.be/en/research-institutes/eli/elim/pr-jacques-mahillon.html J.Mahillon] unpublished), two members of the [[IS Families/IS5 and related IS1182 families|IS''5'' family]], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427'']<ref><pubmed>1963949</pubmed></ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISMk1 IS''Mk1'']<ref><pubmed>8409920</pubmed></ref> and various members of the [[IS Families/IS630 family|IS''630'' family]] including [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS870 IS''870''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISRf1 IS''Rf1'']<ref><pubmed>8387998</pubmed></ref>. Instead, some of these elements insert into a relatively specific target sequence in which the target DR produced on insertion itself generates the Tpase termination codon (see: [[IS Families/IS630 family|IS''630'' family]]). The relevance of this as a control mechanism has yet to be explored. |
===Transposase stability=== | ===Transposase stability=== | ||
| − | Transposase stability can also contribute to the control of transposition activity. The Tpase of IS''903'' is sensitive to the ''E. coli'' Lon protease<ref | + | Transposase stability can also contribute to the control of transposition activity. The Tpase of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] is sensitive to the ''E. coli'' Lon protease<ref name=":6"><pubmed>2161528</pubmed></ref>. This sensitivity limits the activity of the Tpase both temporally and spatially and may provide an explanation for the observation that several Tpases function preferentially in cis. Indeed mutant [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] Tpase derivatives have been isolated which exhibits an increased capacity to function in trans. These are more refractory to Lon degradation than the wildtype protein<ref name=":7"><pubmed>8898394</pubmed></ref>. Some evidence that Lon may also be involved in regulating [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']) transposition has also been presented<ref name=":4" />. |
| − | An observation which might also reflect Tpase instability is the temperature sensitive nature of IS''1''-mediated adjacent deletions ''in vivo''<ref | + | An observation which might also reflect Tpase instability is the temperature sensitive nature of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']-mediated adjacent deletions ''in vivo''<ref><pubmed>1101028</pubmed></ref>, of [https://tncentral.ncc.unesp.br/report/te/Tn3-V00613 Tn''3'' transposition]<ref><pubmed>378977</pubmed></ref> and of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''] intramolecular recombination both ''in vivo'' and ''in vitro''<ref><pubmed>9302015</pubmed></ref>. For [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''], incubation of the Tpase at 42°C results in an irreversible loss in activity. Further analyses showed that there was an increase in the proportion of transposase fragments some of which can presumably interact with full length transposase to inhibit its activity. This effect can be somewhat reduced by a series of mutation in the transposase gene whose function in stabilizing the transposase is as yet unknown<ref><pubmed>17078817</pubmed></ref>. |
===Co-translational binding and multimerization=== | ===Co-translational binding and multimerization=== | ||
| − | [[Image:1.36.1.png|thumb| | + | [[Image:1.36.1.png|thumb|600x600px|'''Fig.21.5.''' Co-translational binding. Left. Artist impression. An RNA polymerase molecule (single blue sphere) in the process of transcribing the transposase gene while the following ribosome (double blue sphere) translates the N-terminal region of the transposase (brown ellipses). The emerging N-terminal ribosome-associated peptide is anchored close to the IS by the mRNA emerging from the RNApol transcription complex. The emerging peptide is shown binding to the neighboring double-strand end IS (green helix). Binding occurs within a short window of time during translation but before the C-terminus of the transposase has been synthesized. The C-terminal end is thought to mask the transposase N-terminal DNA binding domain preventing binding of the full-length protein. This type of control would prevent excessive transposition of other ISs in the cell (and consequent deleterious effects on genome integrity) due to the presence of free active transposase. Artwork adapted by David Villa (components from Fotolia). |
| − | '''Right'''. Cartoon illustrating cotranslational binding. IRL and IRR are indicated as is the indigenous promoter pIRL, located partially in IRL. RNA polymerase (blue), both ribosome sub units (blue) and mRNA are also indicated. The nascent peptide is shown in brown. The cartoon is not to scale.|alt=]]Certain prokaryotic IS transposases show a strong preference for acting on the element from which they are expressed rather than on other copies of the same element in the cell. This phenomenon of “cis” preference presumably serves to prevent general activation of several identical IS copies by any “accidental” (stochastic) transposase expression from a single IS. Several different IS such as IS''1''<ref> | + | '''Right'''. Cartoon illustrating cotranslational binding. IRL and IRR are indicated as is the indigenous promoter pIRL, located partially in IRL. RNA polymerase (blue), both ribosome sub units (blue) and mRNA are also indicated. The nascent peptide is shown in brown. The cartoon is not to scale.|alt=]]Certain prokaryotic IS transposases show a strong preference for acting on the element from which they are expressed rather than on other copies of the same element in the cell. This phenomenon of “cis” preference presumably serves to prevent general activation of several identical IS copies by any “accidental” (stochastic) transposase expression from a single IS. Several different IS such as [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']''<ref name=":2" />'', [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']<ref><pubmed>6299577</pubmed></ref>, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']<ref><pubmed>6291787</pubmed></ref>, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903'']<ref name=":8"><pubmed>6271455</pubmed></ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911'']<ref name=":9"><pubmed>22195971</pubmed> |
| + | </ref> (see <ref><pubmed>15207871</pubmed></ref><ref name=":6" /> and references therein) exhibit this regulatory phenotype but “cis” preference may be the result of a combination of diverse mechanisms. Thus the Lon protease enhances “cis” preference of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposase <ref name=":6" />. Transposition is enhanced in the absence of Lon and can be overcome by increased transposase expression<ref name=":7" />. For [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''], it is influenced by translation levels, Tpase mRNA half-life and translation efficiency<ref><pubmed>7692216</pubmed></ref><ref><pubmed>8412678</pubmed></ref>. | ||
| − | Another mechanism, co-translational binding based on tight coupling between prokaryotic transcription and translation, was proposed to explain the inability to complement a Tpase mutant of IS''903''<ref | + | Another mechanism, co-translational binding based on tight coupling between prokaryotic transcription and translation, was proposed to explain the inability to complement a Tpase mutant of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903'']''<ref name=":8" />'' and, more specifically, for [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']<ref><pubmed>6296834</pubmed></ref>. |
| − | Some full length IS transposases bind weakly to their cognate IR but the isolated DNA binding domain can bind more strongly. This has been observed for transposases of several elements including IS''1''<ref | + | Some full length IS transposases bind weakly to their cognate IR but the isolated DNA binding domain can bind more strongly. This has been observed for transposases of several elements including [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']<ref><pubmed>2826132</pubmed></ref> and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS30 IS''30'']<ref><pubmed>15469518</pubmed></ref><ref><pubmed>2154486</pubmed></ref> and has also been observed for that of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911'']. Early studies using band shift assays demonstrated that full length OrfAB binds the IRs only weakly and that OrfA binding was even lower or undetectable<ref><pubmed>10677279</pubmed></ref><ref><pubmed>11352577</pubmed></ref>. However, a truncated version of OrfAB, OrfAB [1-149], which is amputated for the C-terminal catalytic domain bound both ends avidly<ref><pubmed>9761671</pubmed></ref> (see also "[[General Information/Transposase expression and activity#Transposase stability|Transposase Stability]]"). It is important to note this implies that, in many ''in vitro'' systems, the majority of transposase is thus likely to be inactive or only partially active since it would not bind stably to its substrate. The observations suggest that the C-terminal (C-ter) domain inhibits specific binding by the sequence-specific N-terminal DNA binding domain possibly by steric masking [[:File:1.36.1.png|(Fig.21.5)]]. This idea is consistent with the observation that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] transposase activity is increased by partial denaturation (for example by treatment with low alcohol concentrations; <ref><pubmed>8132525</pubmed></ref>). It is also consistent with the observation that the OrfAB protein of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS2 IS''2''] can bind the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS2 IS''2''] IRs when it carries a large GFP tag<ref><pubmed>22032517</pubmed></ref><ref><pubmed>22277150</pubmed></ref>. |
| − | One biological explanation for cis preference is that the nascent N-ter domain might fold before completion of translation of the C-ter domain and the nascent protein could initiate binding directly to the closest IS end. Once bound, it would no longer be sensitive to masking by the C-ter domain. If binding fails to occur after translation of the N-ter DNA binding domain, continuing translation and folding of the C-ter domain would then mask the DNA binding domain resulting in an inactive protein. This implies that binding necessary for subsequent catalysis would occur only transitorily early in translation [[:File:1.36.1.png|(Fig. | + | One biological explanation for cis preference is that the nascent N-ter domain might fold before completion of translation of the C-ter domain and the nascent protein could initiate binding directly to the closest IS end. Once bound, it would no longer be sensitive to masking by the C-ter domain. If binding fails to occur after translation of the N-ter DNA binding domain, continuing translation and folding of the C-ter domain would then mask the DNA binding domain resulting in an inactive protein. This implies that binding necessary for subsequent catalysis would occur only transitorily early in translation [[:File:1.36.1.png|(Fig.21.5)]]. |
| − | Direct evidence for co-translational binding was provided for IS''911'' using an ''in vitro'' transcription/translation system<ref | + | Direct evidence for co-translational binding was provided for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''] using an ''in vitro'' transcription/translation system<ref name=":9" /> where it was also demonstrated that reducing the efficiency of the -1 translational frameshifting required for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS911 IS''911''] transposase expression resulted in an increase in binding of a nascent transposase peptide. This is presumably because slowing the frameshifting process increases the time that the N-terminal part of the protein (which carries the sequence-specific DNA binding domain) is present on the ribosome enhancing its probability of binding to a neighboring IS end. It is interesting to note that in many IS, the DNA binding domain which recognizes the IR is located at the N-terminal end of the protein which is translated first. |
| − | One of the remaining questions concerns transposase multimerization. They must form multimers within the transpososome at some stage in the transposition pathway. Some transposases are monomeric in the absence of DNA (e.g. MuA and Tn''5''; <ref | + | One of the remaining questions concerns transposase multimerization. They must form multimers within the transpososome at some stage in the transposition pathway. Some transposases are monomeric in the absence of DNA (e.g. [https://www.uniprot.org/uniprot/A7ZK38 MuA] and [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']; <ref><pubmed>1655409</pubmed></ref><ref><pubmed>9867814</pubmed></ref><ref><pubmed>18680433</pubmed></ref>) while others are multimeric dimeric in solution (e.g. INHIV-1; <ref><pubmed>12446721</pubmed></ref>; <ref><pubmed>17157316</pubmed></ref>; <ref><pubmed>19609359</pubmed></ref>; <ref><pubmed>19229293</pubmed></ref>). |
In view of the possibility that many transposases undergo co-translational binding, and the observation that several different purified full-length transposases bind poorly to the ends of their cognate transposon (while the isolated N-terminal DNA binding domain alone binds robustly), it must be emphasized that purified transposases are probably largely inactive. This must be taken into account when assessing transposase properties. | In view of the possibility that many transposases undergo co-translational binding, and the observation that several different purified full-length transposases bind poorly to the ends of their cognate transposon (while the isolated N-terminal DNA binding domain alone binds robustly), it must be emphasized that purified transposases are probably largely inactive. This must be taken into account when assessing transposase properties. | ||
| − | A recent study has provided support for the idea that transposases may also be able to multimerize cotranslationally. This study, has shown that bacterial luciferase subunits LuxA and LuxB may assemble cotranslationally i''n vivo.'' This process requires ribosome-associated trigger factor. This chaperone apparently delays subunit interactions until the LuxB dimer interface is available<ref | + | A recent study has provided support for the idea that transposases may also be able to multimerize cotranslationally. This study, has shown that bacterial [[wikipedia:Luciferase|luciferase]] subunits [https://www.uniprot.org/uniprot/Q91UU4 LuxA] and [https://www.uniprot.org/uniprot/P19840 LuxB] may assemble cotranslationally i''n vivo.'' This process requires ribosome-associated trigger factor. This chaperone apparently delays subunit interactions until the [https://www.uniprot.org/uniprot/P19840 LuxB] dimer interface is available<ref><pubmed>26405228</pubmed></ref>. |
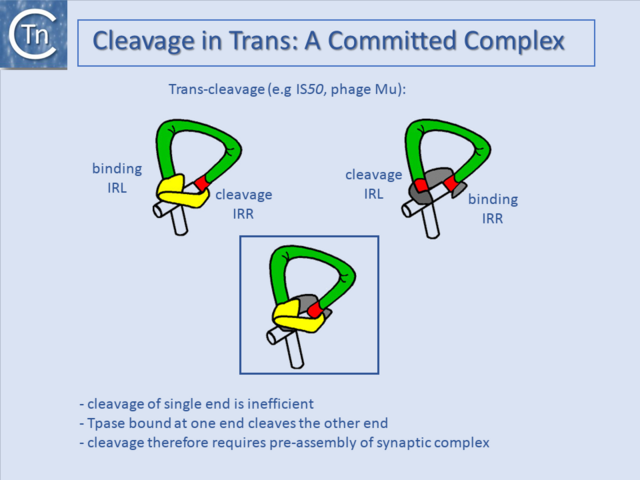
===Cleavage in Trans: A Committed Complex=== | ===Cleavage in Trans: A Committed Complex=== | ||
| − | Another level of regulation often occurs within the transpososome (see [[General Information/Reaction mechanisms|Reaction Mechanisms]]) itself. Transposases bound to their specific recognition sequence, generally at both ends of the TE, form a | + | Another level of regulation often occurs within the transpososome (see "[[General Information/Reaction mechanisms|Reaction Mechanisms]]") itself. Transposases bound to their specific recognition sequence, generally at both ends of the TE, form a '''S'''ynaptic (SC) or '''P'''aired-'''E'''nd '''C'''omplex (PEC) with a precise architecture for activity, and the transposase bound at one end is used to cleave the opposite end ([[:File:1.36.2.png|Fig.21.6]]) (e.g. <ref><pubmed>23135398</pubmed></ref> and <ref><pubmed>10884228</pubmed></ref>). This ensures that the correct SC is formed prior to cleavage and strand transfer and thus avoids the generation of nonproductive cleavage products, which would cause damage to the donor DNA molecule. |
| − | |||
| − | |||
| − | |||
| − | + | [[Image:1.36.2.png|thumb|640x640px|'''Fig.21.6.''' Trans-cleavage. The figure shows a transposase dimer in a paired-end with two transposons ends. Cleavage of single-ends is inefficient. Transposase bound at one end cleaves the other end. Cleavage, therefore, requires preassembly of the synaptic complex.|alt=|center]] | |
| − | |||
===Host factors=== | ===Host factors=== | ||
| − | Transposition activity is frequently modulated by various host factors. These effects are generally specific for each element. A non-exhaustive list of such factors includes the DNA chaperones (or histone-like proteins), IHF, HU, HNS, and FIS, the replication initiator DnaA, the protein chaperone/proteases ClpX, | + | Transposition activity is frequently modulated by various host factors. These effects are generally specific for each element. A non-exhaustive list of such factors includes the DNA chaperones (or [[wikipedia:Bacterial_DNA_binding_protein|histone-like proteins]]), [[wikipedia:Bacterial_DNA_binding_protein#IHF|IHF]], [[wikipedia:Bacterial_DNA_binding_protein#HU|HU]], [[wikipedia:Bacterial_DNA_binding_protein#H-NS|HNS]], and FIS, the replication initiator [[wikipedia:DnaA|DnaA]], the protein chaperone/proteases [[wikipedia:ClpX|ClpX]], [[wikipedia:Clp_protease_family|ClpP]], and [[wikipedia:Clp_protease_family|ClpA]], the [[wikipedia:SOS_response|SOS control]] protein [[wikipedia:Repressor_lexA|LexA]], and the Dam DNA methylase. In addition, proteins which govern DNA supercoiling in the cell can also influence transposition. |
====IHF, HU, HNS, and FIS==== | ====IHF, HU, HNS, and FIS==== | ||
| − | DNA chaperones may play roles in assuring the correct three dimensional architecture in the evolution of various nucleoprotein complexes necessary for productive transposition. They may also be involved in regulating Tpase expression. IHF, HU, HNS, and FIS have all been variously implicated in the case of bacteriophage Mu, in the control of Mu gene expression or directly in the transposition process (see <ref | + | DNA chaperones may play roles in assuring the correct three dimensional architecture in the evolution of various nucleoprotein complexes necessary for productive transposition. They may also be involved in regulating Tpase expression. [[wikipedia:Bacterial_DNA_binding_protein#IHF|IHF]], [[wikipedia:Bacterial_DNA_binding_protein#HU|HU]], [[wikipedia:Bacterial_DNA_binding_protein#H-NS|HNS]], and FIS have all been variously implicated in the case of [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]], in the control of Mu gene expression or directly in the transposition process (see <ref><pubmed>8805293</pubmed></ref> for review). |
| − | Several elements carry specific binding sites for IHF within, or close to, their terminal IRs. These can lie within (e.g. IS''1'': <ref | + | Several elements carry specific binding sites for IHF within, or close to, their terminal IRs. These can lie within (e.g. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']: <ref><pubmed>2995832</pubmed></ref>; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903'']: see <ref><pubmed>6261245</pubmed></ref> or close to ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] <ref name=":1" />) the Tpase promoter. [[wikipedia:Bacterial_DNA_binding_protein#IHF|IHF]] appears to influence the nature of IS''10'' transposition products by binding to a site 43 bp from one end <ref><pubmed>7744253</pubmed></ref><ref name=":10"><pubmed>9630232</pubmed> |
| + | </ref><ref><pubmed>7556079</pubmed></ref>. | ||
| − | IHF also stimulates Tpase binding to the ends of the Tn''3'' family member, Tn''1000'' or γδ <ref | + | IHF also stimulates Tpase binding to the ends of the [[Transposons families/Tn3 family|Tn''3'' family]] member, [https://tncentral.ncc.unesp.br/report/te/Tn1000-X60200.1 Tn''1000''] or γδ <ref><pubmed>2844529</pubmed></ref>. Ironically, although [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] was the first element in which IHF sites were identified (one within each IR), conditions have not yet been found in which IHF shows a clear effect on transposition or gene expression (D. Zerbib and [https://scholar.google.com/citations?user=r8TYgVEAAAAJ&hl=pt-BR M. Chandler], unpublished results). |
| − | The multiple roles of several of these proteins in both the IS''10'' and Tn''5'' (IS''50'') systems and the dynamics of their involvement has been determined in detail<ref | + | The multiple roles of several of these proteins in both the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']) systems and the dynamics of their involvement has been determined in detail<ref name=":10" /><ref><pubmed>11447129</pubmed></ref><ref><pubmed>14530435</pubmed></ref><ref><pubmed>14992723</pubmed></ref><ref name=":11"><pubmed>19696075</pubmed> |
| + | </ref><ref><pubmed>19042975</pubmed></ref><ref name=":12"><pubmed>16166383</pubmed></ref><ref name=":13"><pubmed>17501923</pubmed></ref>. | ||
| − | The histone-like nucleoid structuring protein H-NS, a global transcriptional regulator, has also been implicated in the regulation of bacterial transposition systems, including Tn''10''<ref | + | The histone-like nucleoid structuring protein H-NS, a global transcriptional regulator, has also been implicated in the regulation of bacterial transposition systems, including [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10'']''<ref name=":11" />''<ref name=":12" /><ref name=":13" />. It appears to promote transposition by binding directly to the transposition complex (or transpososome). |
====DnaA==== | ====DnaA==== | ||
| − | In the case of IS''50'', an element of the same family as IS''10'', both the protein Fis and the replication initiator protein DnaA have been reported to intervene in transposition (see <ref | + | In the case of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''], an element of the same family as [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''], both the protein Fis and the replication initiator protein [[wikipedia:DnaA|DnaA]] have been reported to intervene in transposition (see <ref><pubmed>7504907</pubmed></ref>). Finally another "histone-like" protein, HNS, has been reported to stimulate transposition of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] in certain circumstances<ref><pubmed>10583504</pubmed></ref>. |
Although their mode of action is at present unknown, several other host proteins with otherwise entirely different functions have been implicated in transposition. | Although their mode of action is at present unknown, several other host proteins with otherwise entirely different functions have been implicated in transposition. | ||
| − | Accessory proteins: Acyl carrier protein (ACP), ribosomal protein L29, PepA and ArgR | + | Accessory proteins: [[wikipedia:Acyl_carrier_protein|Acyl carrier protein]] (ACP), ribosomal protein L29, PepA and ArgR |
| − | Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of Tn''3'' by its cognate Tpase<ref | + | Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of [https://tncentral.ncc.unesp.br/report/te/Tn3-V00613 Tn''3''] by its cognate Tpase<ref name=":17"><pubmed>9077464</pubmed></ref> and, together with [[wikipedia:60S_ribosomal_protein_L29|ribosomal protein L29]], to greatly increase binding of TnsD (a protein involved in [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] target selection) to the chromosomal insertion site, attTn7 <ref name=":12" />. Moreover ACP and L29 moderately stimulate [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] transposition ''in vitro'' while L29 alone has a significant stimulatory effect ''in vivo <ref name=":12" />''. The mode of action of these proteins may be similar to that of the accessory proteins [https://www.uniprot.org/uniprot/P68767 PepA] and [https://www.uniprot.org/uniprot/P0A6D0 ArgR] which modify the architecture of the synaptic complex in certain [[wikipedia:Site-specific_recombination|XerC/XerD-mediated site-specific recombination]] reactions<ref name=":14"><pubmed>9348666</pubmed></ref>. |
| − | <br /> | + | |
| + | ====Accessory proteins: Acyl carrier protein (ACP), ribosomal protein L29, PepA and ArgR==== | ||
| + | Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of [https://tncentral.ncc.unesp.br/report/te/Tn3-V00613 Tn''3''] by its cognate Tpase <ref name=":17" /> and, together with ribosomal protein [[wikipedia:60S_ribosomal_protein_L29|L29]], to greatly increase binding of TnsD (a protein involved in [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] target selection) to the chromosomal insertion site, ''attTn7'' <ref name=":15"><pubmed>9755182</pubmed></ref>. Moreover ACP and [[wikipedia:60S_ribosomal_protein_L29|L29]] moderately stimulate [https://tncentral.ncc.unesp.br/report/te/Tn7-NC_002525 Tn''7''] transposition in vitro while [[wikipedia:60S_ribosomal_protein_L29|L29]] alone has a significant stimulatory effect ''in vivo'' <ref name=":15" />. The mode of action of these proteins may be similar to that of the accessory proteins [https://www.uniprot.org/uniprot/P68767 PepA] and [https://www.uniprot.org/uniprot/P0A6D0 ArgR] which modify the architecture of the synaptic complex in certain [[wikipedia:Site-specific_recombination|XerC/XerD-mediated site-specific recombination]] reactions <ref name=":14" />. | ||
| + | ====ClpX, ClpP, and Lon==== | ||
| + | '''<big>C</big>'''ertain factors involved in protein "management" such as [[wikipedia:ClpX|ClpX]], [[wikipedia:Clp_protease_family|ClpP]], and [[wikipedia:Lon_protease_family|Lon]] have been implicated in transposition. [[wikipedia:ClpX|ClpX]] is essential for [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]] growth<ref><pubmed>8022280</pubmed></ref> where it is required for disassembling the transposase-DNA complex or the transpososome strand transfer complex in preparation for the assembly of a replication complex<ref><pubmed>8631314</pubmed></ref><ref><pubmed>7557391</pubmed></ref>. Recognition of [https://www.uniprot.org/uniprot/P07636 Mu transposase], pA, by [[wikipedia:ClpX|ClpX]] requires the terminal 10 amino acids of [https://www.uniprot.org/uniprot/P07636 pA]<ref><pubmed>9203582</pubmed></ref>. Together with [[wikipedia:Clp_protease_family|ClpP]], [[wikipedia:ClpX|ClpX]] also plays a role in proteolysis of the [https://www.uniprot.org/uniprot/P06019 Mu repressor]<ref><pubmed>8617219</pubmed></ref><ref><pubmed>9299335</pubmed></ref>. The [[wikipedia:Lon_protease_family|Lon]] protease is implicated in proteolysis of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposase <ref name=":6" />. | ||
| + | |||
| + | At present the involvement of these proteins in the transposition of other elements has not been well documented. | ||
| + | ====SOS system, RecA, RecBC==== | ||
| + | The third class of host factor includes host cell systems which act to limit DNA damage and maintain chromosome integrity. Studies with [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] (see <ref name=":1" />) and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']<ref><pubmed>7932694</pubmed></ref> have demonstrated that high levels of Tpase in the presence of suitable terminal IRs lead to induction of the host [[wikipedia:SOS_response|SOS system]]. As discussed previously<ref><pubmed>9729608</pubmed></ref>, some controversy still exists concerning the role of [[wikipedia:RecA|RecA]] in [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50'']) transposition<ref name=":16"><pubmed>1648004</pubmed></ref><ref name=":22"><pubmed>1655708</pubmed></ref><ref name=":32"><pubmed>1657870</pubmed></ref>. [https://biochem.wisc.edu/emeritus/reznikoff/cv Reznikoff] and colleagues have provided genetic evidence that transposition is inhibited by induction of the [[wikipedia:SOS_response|SOS system]] in a manner which does not require the proteolytic activity of [[wikipedia:RecA|RecA]]<ref name=":32" />. On the other hand, Tessman and collaborators<ref name=":16" /><ref name=":22" /><ref><pubmed>1328165</pubmed></ref> using a different transposition assay have found that constitutive [[wikipedia:SOS_response|SOS]] conditions actually enhance [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] transposition. Moreover, using yet another assay system, Ahmed<ref name=":02"><pubmed>3025455</pubmed> | ||
| + | </ref> has concluded that intermolecular transposition of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] is stimulated by [https://www.uniprot.org/uniprot/P0A7G6 RecA]. Further investigation is clearly required to understand these apparently incompatible results. | ||
| + | |||
| + | Ahmed has also concluded that intermolecular transposition of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1'']-based transposon, Tn''9'', behaves in a similar way to that of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5''] with respect to the ''[[wikipedia:RecA|recA]]'' allele<ref name=":02" />. In contrast, however, the frequency of adjacent deletions mediated by [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] was significantly increased in the absence of [[wikipedia:RecA|RecA]]. This has received some independent support using a physical assay where it was shown that deletion products accumulate in a ''[[wikipedia:RecA|recA]]'' but not in a wildtype host. Moreover, like [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''] induction of the [[wikipedia:SOS_response|SOS system]], accumulation of such adjacent deletions was dependent on ''recBC'' (Zablweska et al., unpublished observations). The ''recBC'' genes are also implicated in the behavior of transposons such as [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] and [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']<ref name=":02" /> where they affect precise and imprecise excision in a process independent of transposition per se. This is more pronounced with composite transposons in which the component insertion sequences [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''] are present as inverted repeats, and is stimulated when the transposon is carried by a transfer-proficient conjugative plasmid. It seems probable that such excisions occur by a process involving replication fork slippage (see <ref><pubmed>10844241</pubmed></ref><ref><pubmed>10715006</pubmed></ref> for further discussion). | ||
| + | ====PolI and gyrase==== | ||
| + | Both [[wikipedia:DNA_polymerase_I|DNA polymerase I]] <ref><pubmed>6267432</pubmed></ref><ref><pubmed>6288966</pubmed></ref><ref><pubmed>3030303</pubmed></ref> and [[wikipedia:DNA_gyrase|DNA gyrase]]<ref><pubmed>6290084</pubmed></ref><ref><pubmed>6265907</pubmed></ref> are implicated in the transposition of [https://tncentral.ncc.unesp.br/report/te/Tn5-U00004.1 Tn''5'']. While the effect of gyrase may reflect a requirement for optimal levels of supercoiling, the role of [[wikipedia:DNA_polymerase_I|PolI]] remains a matter of speculation. It may be involved in DNA synthesis necessary to repair the single strand gaps resulting from staggered cleavage of the target and which gives rise to the DRs. DNA gyrase has also been shown to be important in transposition of [[wikipedia:Bacteriophage_Mu|bacteriophage Mu]]<ref><pubmed>7592347</pubmed></ref><ref><pubmed>8930913</pubmed></ref>. | ||
| + | ====Dam methylase==== | ||
| + | Another host function, the [[wikipedia:DNA_adenine_methylase|Dam DNA methylase]] can be important in modulating both Tpase expression and activity. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] all carry methylation sites (GATC) in the transposase promoter regions and in each case, promoter activity is increased in a dam-host<ref name=":42"><pubmed>3000598</pubmed></ref><ref><pubmed>2451025</pubmed></ref>. Additional evidence has been presented that the methylation status of GATC sites within the terminal inverted repeats also modulates the activity of these ends<ref name=":42" />. For [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''], this can now be understood in terms of steric interference in the transposase active site, as recently revealed by the determination of the crystal structure of a synpatic complex including its Tpase and a pair of precleaved transposon ends<ref><pubmed>10207011</pubmed></ref>. Similar methylation sites have been previously observed in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS3 IS''3''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS4 IS''4''], and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5'']. A survey of the elements included in the data base has shown that most groups or families contain members which have GATC sites within the first 50 bp of one or both extremities. The [[IS Families/IS3 family|IS''3'']]'','' [[IS Families/IS5 and related IS1182 families|IS''5'']] and [[IS Families/IS256 family|IS''256'']] families include the most members carrying such sites. Except for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS3 IS''3''] itself where strong stimulation of transposition has been observed in a dam-host, in most of these cases the biological relevance of these sites is unknown. Moreover, it should be pointed out that the probability that any 100 bp DNA sequence carries the GATC tetranucleotide is about 40%. The role of [[wikipedia:DNA_adenine_methylase|Dam methylation]] in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS50R IS''50''] transposition is described in detail in the appropriate sections dealing with these elements. | ||
| + | ====Metabolic control elements==== | ||
| + | In a screen of over 20,000 independent insertion mutants for host factors that influence [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposition the [https://www.derbylab.org/ Derbyshire lab] isolated more than 100 mutants that increased or decreased transposition and also altered its timing during colony growth<ref><pubmed>16135227</pubmed></ref>. These included independent mutations in a gene required for fermentative metabolism during anaerobic growth resulting in “early” transposition during colony growth and was suppressed by addition of fumarate, and other mutations in genes associated with DNA metabolism, intermediary metabolism, transport, cellular redox, protein folding and proteolysis. Other mutations were isolated in pur genes involved in purine biosynthesis. Further analysis suggested that this phenotype was due to a requirement for GTP in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposition<ref><pubmed>15968071</pubmed></ref>. It should be noted that some of these mutants also affected transposition of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and of Tn''552''. | ||
| + | ====Hfq==== | ||
| + | Finally, the RNA chaperone [[wikipedia:Hfq_protein|Hfq]] has also been implicated in the regulation of [https://tncentral.ncc.unesp.br/report/te/Tn10-AF162223 Tn''10''] transposition by promoting RNAout interaction with transposase mRNA<ref><pubmed>23510801</pubmed></ref><ref><pubmed>25649688</pubmed></ref><ref><pubmed>25579599</pubmed></ref>. There is also some suggestion that SmAP1, the archaeal Hfq homolog, may also play a role in regulation of transposition of several different archaeal IS <ref><pubmed>36912639</pubmed></ref> but this remains to be tested directly. | ||
| + | ===Over-production inhibition=== | ||
| + | Certain transposons appear to be subject to a mode of regulation known as over-expression inhibition. This was first observed with the eukaryotic transposons Tc1/mariner Lampe<ref><pubmed>9584095</pubmed></ref><ref><pubmed>8882498</pubmed></ref><ref><pubmed>9154003</pubmed></ref> where increasing the concentration of transposase results in a reduction in the level of transposition. It was subsequently observed with the sleeping beauty transposon<ref><pubmed>12842434</pubmed></ref><ref><pubmed>14759813</pubmed></ref>. It also occurs ''in vivo'' in mice<ref><pubmed>14961361</pubmed></ref><ref><pubmed>14529839</pubmed></ref>. | ||
| + | The biological rational for this is that “infection” of a naïve cell by the transposon results in a burst of transposition which is then attenuated by overproduction inhibition. This is then followed by gradual decay of the transposon. | ||
| + | The [https://chalmerslab.wordpress.com/research/ Chalmers lab]<ref><pubmed>23795293</pubmed></ref> has provided an interesting and compelling explanation of this effect. Using the mariner family transposon Hsmar1 they present convincing data implying that overproduction inhibition occurs during transpososome assembly and is due to a combination of the multimeric state of the transposase coupled with competition for transposase binding sites at the Hsmar1 ends<ref><pubmed>24812590</pubmed></ref><ref><pubmed>26104691</pubmed></ref>. The model (assembly-site-occlusion model) is based on the presence of transposase multimers (dimers) to the exclusion of monomers – in other words, end-binding required a dimeric transposase. At low transposase/transposon ratios, one dimer can bind both transposon ends resulting in the ordered assembly of the transpososome. An increase in the transposase dimer/transposon ratio results in binding of dimers to both transposon ends, preventing transpososome assembly. The model not only explains the ''in vivo'' transposase dose-response for Hsmar1 but also for the related Sleeping Beauty (SB) and piggyBac (PB) transposons. As yet, no information is at present available concerning the relevance of this mode of regulation to prokaryotic transposable elements. | ||
==Bibliography== | ==Bibliography== | ||
| − | < | + | {{Reflist|32em}} |
| + | <br /> | ||
| + | <hr> | ||
| + | {{TnPedia}} | ||
Latest revision as of 05:07, 1 April 2024
Contents
- 1 Transposase expression and activity
- 1.1 Impinging transcription
- 1.2 Sequestration of translation initiation signals
- 1.3 Programmed Translational Frameshifting
- 1.4 Programmed Transcriptional Frameshifting
- 1.5 Recoding suppression of stop codons using the unusual amino acids Pyrrolysine and Selenocysteine
- 1.6 Translation termination
- 1.7 Transposase stability
- 1.8 Co-translational binding and multimerization
- 1.9 Cleavage in Trans: A Committed Complex
- 1.10 Host factors
- 1.11 Over-production inhibition
- 2 Bibliography
Transposase expression and activity
While many of the classical mechanisms of controlling gene expression, such as the production of transcriptional repressors (IS1: [1][2]; IS2: [3] or translational inhibitors (anti-sense RNA in the case of IS10; see [4] are known to operate in Tpase expression, several other mechanisms have also been uncovered.
Impinging transcription
Many ISs have evolved mechanisms which attenuate their activation by impinging transcription following insertion into active host genes. This was originally observed in the case of IS1[5][6], for bacteriophage Mu[7] and IS50. To our knowledge other elements have not been examined.
This effect may be the result of disrupting complexes between Tpase and cognate DNA ends and could reflect either inhibition of transposase binding or disruption of extant transposase-end complexes.
In the case of bacteriophage Mu, transcription originating from within the element and impinging on the left end has also been shown to reduce activity[7]. It is possible that transcription disrupts the formation of intermediates including transposase and one or both Mu ends which lead to stable transposition complexes.
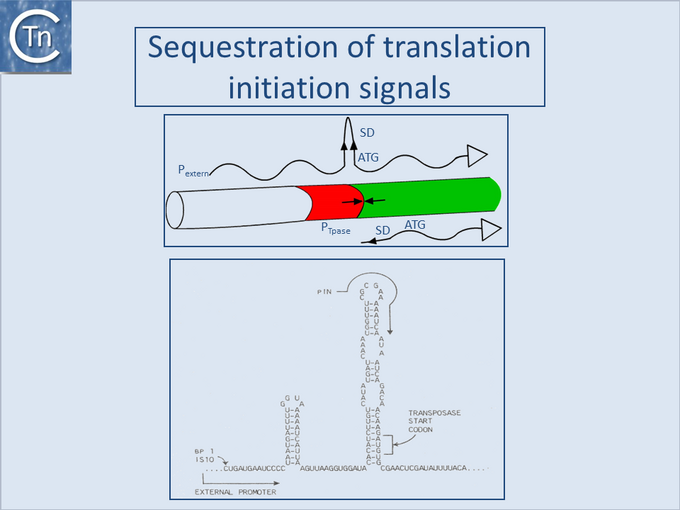
Sequestration of translation initiation signals
One such mechanism observed with IS10 and IS50, and potentially present in several other ISs, is the sequestering of translation initiation signals in an RNA secondary structure (Fig.21.1). These ISs carry inverted repeat sequences located close to the left end which include the ribosome binding site or translation initiation codon for the Tpase gene. Transcripts from the resident Tpase promoter include only the distal repeat unit which is unable to form the secondary structure, while transcripts from neighboring DNA include both repeats and would generate secondary structures in the mRNA which would sequester translation initiation signals[8][9]. This has been demonstrated experimentally for IS10 and IS50 but several additional insertion sequences carry such potential structures and might be expected to exhibit a similar mechanism (see [10]).
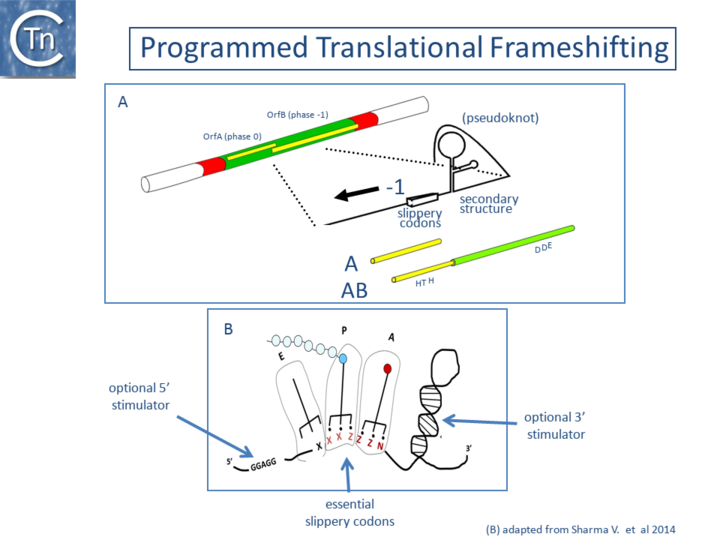
Programmed Translational Frameshifting
A second mechanism acts at the level of translation elongation and involves programmed translational frameshifting between two consecutive open reading frames (Fig.21.1). Typically a -1 frameshift is observed in which the translating ribosome slides one base upstream and resumes in the alternative phase. This generally occurs at the position of so-called slippery codons in a heptanucleotide sequence of the type X XXZ ZZN in phase 0 (where the bases paired with the anticodon are shown as triplets) which is read as XXX ZZZ N in the shifted -1 phase (Fig.21.2) (see e.g. [11][12][13], http://recode.genetics.utah.edu/). The sequence A AAA AAG is a common example of this type of heptanucleotide. Ribosomal shifting of this type is stimulated by structures in the mRNA which tend to impede the progression of the ribosome such as potential ribosome binding sites upstream or secondary structures (stem-loop structures and pseudoknots) downstream of the slippery codons. Translational control of transposition by frameshifting has been demonstrated both for IS1[14][2], and for members of the IS3 family (Fig.21.3) ([15]; see also [11]) but may also occur in several other IS elements (see for example IS5 and IS630 families). For IS1 and members of the IS3 family, the upstream frame appears to carry a DNA recognition domain whereas the downstream frame encodes the catalytic site. While the product of the upstream frame alone acts as a modulator of activity, presumably by binding to the IR sequences, frameshifting assembles both domains into a single protein, the Tpase, which directs the cleavages and strand transfer necessary for mobility of the element. The frameshifting frequency is thus critical in determining overall transposition activity. Although it has yet to be explored in detail, frameshifting could be influenced by host physiology thus coupling transposition activity to the state of the host cell.
Programmed Transcriptional Frameshifting
Another type of frameshifting may also occur in bacterial insertion sequences. This occurs at the transcriptional level and involves misreading and slippage of RNA polymerase on stretches of A residues. Although no real data is at present available, it may occur relatively frequently [16][17][18].
Recoding suppression of stop codons using the unusual amino acids Pyrrolysine and Selenocysteine
Another type of recoding which appears to occur in Methanosarcina, is the “suppression” of the stop codon, UAG, by insertion of Pyrrolysine. This was first noted in the Methylamine methyltransferases which are important in the production of methane by archaeal methanogens [19] identified an in-frame amber codon (TAG) in the trimethylamine methyltransferase genes of both M. barkeri and M. thermophila. However, at least in the case of M. barkeri, abundant quantities of the full-length protein could be obtained and it appeared that the TAG codon was read as Lys. This later proved to be the unusual amino acid pyrrolysine. Several IS copies in these archaeal methanogens carry TAG codons which are presumably “suppressed” by decoding as pyrrolysine.
In this framework, other stop codons are known to be suppressed by decoding as selenocysteine [20][21][22][23]. To our knowledge a single IS from the IS3 family, ISDvu3 from Desulfovibrio vulgaris, includes a selenocysteine inserted at a stop codon in its orfB frame (M. Land personal communication). Undoubtedly additional examples will be identified in the future.
The inclusion of these amino acids involves the presence of specific types of secondary structures in the mRNA (Fig.21.4).
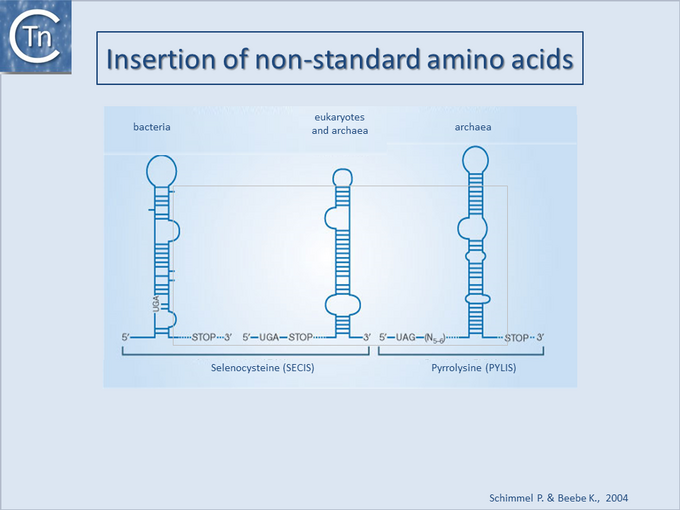
Translation termination
A third potential mechanism derives from the observation that the translation termination codon of Tpase genes of certain elements is located within their IR sequences. Although to our knowledge, no extensive analysis of the significance of this arrangement has yet been undertaken, it seems possible that it may in some manner couple translation termination, transposase binding, and transposition activity. The transposase gene of several elements does not possess a termination codon. These include IS240C, a member of the IS6 family (Y. Chen and J.Mahillon unpublished), two members of the IS5 family, IS427[24] and ISMk1[25] and various members of the IS630 family including IS870 and ISRf1[26]. Instead, some of these elements insert into a relatively specific target sequence in which the target DR produced on insertion itself generates the Tpase termination codon (see: IS630 family). The relevance of this as a control mechanism has yet to be explored.
Transposase stability
Transposase stability can also contribute to the control of transposition activity. The Tpase of IS903 is sensitive to the E. coli Lon protease[27]. This sensitivity limits the activity of the Tpase both temporally and spatially and may provide an explanation for the observation that several Tpases function preferentially in cis. Indeed mutant IS903 Tpase derivatives have been isolated which exhibits an increased capacity to function in trans. These are more refractory to Lon degradation than the wildtype protein[28]. Some evidence that Lon may also be involved in regulating Tn5 (IS50) transposition has also been presented[9].
An observation which might also reflect Tpase instability is the temperature sensitive nature of IS1-mediated adjacent deletions in vivo[29], of Tn3 transposition[30] and of IS911 intramolecular recombination both in vivo and in vitro[31]. For IS911, incubation of the Tpase at 42°C results in an irreversible loss in activity. Further analyses showed that there was an increase in the proportion of transposase fragments some of which can presumably interact with full length transposase to inhibit its activity. This effect can be somewhat reduced by a series of mutation in the transposase gene whose function in stabilizing the transposase is as yet unknown[32].
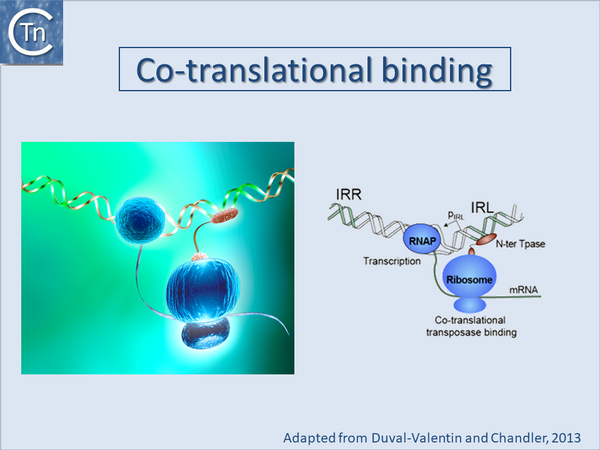
Co-translational binding and multimerization
Certain prokaryotic IS transposases show a strong preference for acting on the element from which they are expressed rather than on other copies of the same element in the cell. This phenomenon of “cis” preference presumably serves to prevent general activation of several identical IS copies by any “accidental” (stochastic) transposase expression from a single IS. Several different IS such as IS1[5], IS10[33], IS50[34], IS903[35] and IS911[36] (see [37][27] and references therein) exhibit this regulatory phenotype but “cis” preference may be the result of a combination of diverse mechanisms. Thus the Lon protease enhances “cis” preference of the IS903 transposase [27]. Transposition is enhanced in the absence of Lon and can be overcome by increased transposase expression[28]. For IS10, it is influenced by translation levels, Tpase mRNA half-life and translation efficiency[38][39].
Another mechanism, co-translational binding based on tight coupling between prokaryotic transcription and translation, was proposed to explain the inability to complement a Tpase mutant of IS903[35] and, more specifically, for Tn5[40].
Some full length IS transposases bind weakly to their cognate IR but the isolated DNA binding domain can bind more strongly. This has been observed for transposases of several elements including IS1[41] and IS30[42][43] and has also been observed for that of IS911. Early studies using band shift assays demonstrated that full length OrfAB binds the IRs only weakly and that OrfA binding was even lower or undetectable[44][45]. However, a truncated version of OrfAB, OrfAB [1-149], which is amputated for the C-terminal catalytic domain bound both ends avidly[46] (see also "Transposase Stability"). It is important to note this implies that, in many in vitro systems, the majority of transposase is thus likely to be inactive or only partially active since it would not bind stably to its substrate. The observations suggest that the C-terminal (C-ter) domain inhibits specific binding by the sequence-specific N-terminal DNA binding domain possibly by steric masking (Fig.21.5). This idea is consistent with the observation that IS10 transposase activity is increased by partial denaturation (for example by treatment with low alcohol concentrations; [47]). It is also consistent with the observation that the OrfAB protein of IS2 can bind the IS2 IRs when it carries a large GFP tag[48][49].
One biological explanation for cis preference is that the nascent N-ter domain might fold before completion of translation of the C-ter domain and the nascent protein could initiate binding directly to the closest IS end. Once bound, it would no longer be sensitive to masking by the C-ter domain. If binding fails to occur after translation of the N-ter DNA binding domain, continuing translation and folding of the C-ter domain would then mask the DNA binding domain resulting in an inactive protein. This implies that binding necessary for subsequent catalysis would occur only transitorily early in translation (Fig.21.5).
Direct evidence for co-translational binding was provided for IS911 using an in vitro transcription/translation system[36] where it was also demonstrated that reducing the efficiency of the -1 translational frameshifting required for IS911 transposase expression resulted in an increase in binding of a nascent transposase peptide. This is presumably because slowing the frameshifting process increases the time that the N-terminal part of the protein (which carries the sequence-specific DNA binding domain) is present on the ribosome enhancing its probability of binding to a neighboring IS end. It is interesting to note that in many IS, the DNA binding domain which recognizes the IR is located at the N-terminal end of the protein which is translated first.
One of the remaining questions concerns transposase multimerization. They must form multimers within the transpososome at some stage in the transposition pathway. Some transposases are monomeric in the absence of DNA (e.g. MuA and Tn5; [50][51][52]) while others are multimeric dimeric in solution (e.g. INHIV-1; [53]; [54]; [55]; [56]).
In view of the possibility that many transposases undergo co-translational binding, and the observation that several different purified full-length transposases bind poorly to the ends of their cognate transposon (while the isolated N-terminal DNA binding domain alone binds robustly), it must be emphasized that purified transposases are probably largely inactive. This must be taken into account when assessing transposase properties.
A recent study has provided support for the idea that transposases may also be able to multimerize cotranslationally. This study, has shown that bacterial luciferase subunits LuxA and LuxB may assemble cotranslationally in vivo. This process requires ribosome-associated trigger factor. This chaperone apparently delays subunit interactions until the LuxB dimer interface is available[57].
Cleavage in Trans: A Committed Complex
Another level of regulation often occurs within the transpososome (see "Reaction Mechanisms") itself. Transposases bound to their specific recognition sequence, generally at both ends of the TE, form a Synaptic (SC) or Paired-End Complex (PEC) with a precise architecture for activity, and the transposase bound at one end is used to cleave the opposite end (Fig.21.6) (e.g. [58] and [59]). This ensures that the correct SC is formed prior to cleavage and strand transfer and thus avoids the generation of nonproductive cleavage products, which would cause damage to the donor DNA molecule.
Host factors
Transposition activity is frequently modulated by various host factors. These effects are generally specific for each element. A non-exhaustive list of such factors includes the DNA chaperones (or histone-like proteins), IHF, HU, HNS, and FIS, the replication initiator DnaA, the protein chaperone/proteases ClpX, ClpP, and ClpA, the SOS control protein LexA, and the Dam DNA methylase. In addition, proteins which govern DNA supercoiling in the cell can also influence transposition.
IHF, HU, HNS, and FIS
DNA chaperones may play roles in assuring the correct three dimensional architecture in the evolution of various nucleoprotein complexes necessary for productive transposition. They may also be involved in regulating Tpase expression. IHF, HU, HNS, and FIS have all been variously implicated in the case of bacteriophage Mu, in the control of Mu gene expression or directly in the transposition process (see [60] for review).
Several elements carry specific binding sites for IHF within, or close to, their terminal IRs. These can lie within (e.g. IS1: [61]; IS903: see [62] or close to (IS10 [4]) the Tpase promoter. IHF appears to influence the nature of IS10 transposition products by binding to a site 43 bp from one end [63][64][65].
IHF also stimulates Tpase binding to the ends of the Tn3 family member, Tn1000 or γδ [66]. Ironically, although IS1 was the first element in which IHF sites were identified (one within each IR), conditions have not yet been found in which IHF shows a clear effect on transposition or gene expression (D. Zerbib and M. Chandler, unpublished results).
The multiple roles of several of these proteins in both the IS10 and Tn5 (IS50) systems and the dynamics of their involvement has been determined in detail[64][67][68][69][70][71][72][73].
The histone-like nucleoid structuring protein H-NS, a global transcriptional regulator, has also been implicated in the regulation of bacterial transposition systems, including Tn10[70][72][73]. It appears to promote transposition by binding directly to the transposition complex (or transpososome).
DnaA
In the case of IS50, an element of the same family as IS10, both the protein Fis and the replication initiator protein DnaA have been reported to intervene in transposition (see [74]). Finally another "histone-like" protein, HNS, has been reported to stimulate transposition of IS1 in certain circumstances[75].
Although their mode of action is at present unknown, several other host proteins with otherwise entirely different functions have been implicated in transposition.
Accessory proteins: Acyl carrier protein (ACP), ribosomal protein L29, PepA and ArgR Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of Tn3 by its cognate Tpase[76] and, together with ribosomal protein L29, to greatly increase binding of TnsD (a protein involved in Tn7 target selection) to the chromosomal insertion site, attTn7 [72]. Moreover ACP and L29 moderately stimulate Tn7 transposition in vitro while L29 alone has a significant stimulatory effect in vivo [72]. The mode of action of these proteins may be similar to that of the accessory proteins PepA and ArgR which modify the architecture of the synaptic complex in certain XerC/XerD-mediated site-specific recombination reactions[77].
Accessory proteins: Acyl carrier protein (ACP), ribosomal protein L29, PepA and ArgR
Acyl carrier protein (ACP) was independently shown to stimulate 3' end cleavage of Tn3 by its cognate Tpase [76] and, together with ribosomal protein L29, to greatly increase binding of TnsD (a protein involved in Tn7 target selection) to the chromosomal insertion site, attTn7 [78]. Moreover ACP and L29 moderately stimulate Tn7 transposition in vitro while L29 alone has a significant stimulatory effect in vivo [78]. The mode of action of these proteins may be similar to that of the accessory proteins PepA and ArgR which modify the architecture of the synaptic complex in certain XerC/XerD-mediated site-specific recombination reactions [77].
ClpX, ClpP, and Lon
Certain factors involved in protein "management" such as ClpX, ClpP, and Lon have been implicated in transposition. ClpX is essential for bacteriophage Mu growth[79] where it is required for disassembling the transposase-DNA complex or the transpososome strand transfer complex in preparation for the assembly of a replication complex[80][81]. Recognition of Mu transposase, pA, by ClpX requires the terminal 10 amino acids of pA[82]. Together with ClpP, ClpX also plays a role in proteolysis of the Mu repressor[83][84]. The Lon protease is implicated in proteolysis of the IS903 transposase [27].
At present the involvement of these proteins in the transposition of other elements has not been well documented.
SOS system, RecA, RecBC
The third class of host factor includes host cell systems which act to limit DNA damage and maintain chromosome integrity. Studies with IS10 (see [4]) and IS1[85] have demonstrated that high levels of Tpase in the presence of suitable terminal IRs lead to induction of the host SOS system. As discussed previously[86], some controversy still exists concerning the role of RecA in Tn5 (IS50) transposition[87][88][89]. Reznikoff and colleagues have provided genetic evidence that transposition is inhibited by induction of the SOS system in a manner which does not require the proteolytic activity of RecA[89]. On the other hand, Tessman and collaborators[87][88][90] using a different transposition assay have found that constitutive SOS conditions actually enhance Tn5 transposition. Moreover, using yet another assay system, Ahmed[91] has concluded that intermolecular transposition of Tn5 is stimulated by RecA. Further investigation is clearly required to understand these apparently incompatible results.
Ahmed has also concluded that intermolecular transposition of the IS1-based transposon, Tn9, behaves in a similar way to that of Tn5 with respect to the recA allele[91]. In contrast, however, the frequency of adjacent deletions mediated by IS1 was significantly increased in the absence of RecA. This has received some independent support using a physical assay where it was shown that deletion products accumulate in a recA but not in a wildtype host. Moreover, like IS1 induction of the SOS system, accumulation of such adjacent deletions was dependent on recBC (Zablweska et al., unpublished observations). The recBC genes are also implicated in the behavior of transposons such as Tn10 and Tn5[91] where they affect precise and imprecise excision in a process independent of transposition per se. This is more pronounced with composite transposons in which the component insertion sequences IS10 and IS50 are present as inverted repeats, and is stimulated when the transposon is carried by a transfer-proficient conjugative plasmid. It seems probable that such excisions occur by a process involving replication fork slippage (see [92][93] for further discussion).
PolI and gyrase
Both DNA polymerase I [94][95][96] and DNA gyrase[97][98] are implicated in the transposition of Tn5. While the effect of gyrase may reflect a requirement for optimal levels of supercoiling, the role of PolI remains a matter of speculation. It may be involved in DNA synthesis necessary to repair the single strand gaps resulting from staggered cleavage of the target and which gives rise to the DRs. DNA gyrase has also been shown to be important in transposition of bacteriophage Mu[99][100].
Dam methylase
Another host function, the Dam DNA methylase can be important in modulating both Tpase expression and activity. IS10, IS50 and IS903 all carry methylation sites (GATC) in the transposase promoter regions and in each case, promoter activity is increased in a dam-host[101][102]. Additional evidence has been presented that the methylation status of GATC sites within the terminal inverted repeats also modulates the activity of these ends[101]. For IS50, this can now be understood in terms of steric interference in the transposase active site, as recently revealed by the determination of the crystal structure of a synpatic complex including its Tpase and a pair of precleaved transposon ends[103]. Similar methylation sites have been previously observed in IS3, IS4, and IS5. A survey of the elements included in the data base has shown that most groups or families contain members which have GATC sites within the first 50 bp of one or both extremities. The IS3, IS5 and IS256 families include the most members carrying such sites. Except for IS3 itself where strong stimulation of transposition has been observed in a dam-host, in most of these cases the biological relevance of these sites is unknown. Moreover, it should be pointed out that the probability that any 100 bp DNA sequence carries the GATC tetranucleotide is about 40%. The role of Dam methylation in IS10 and IS50 transposition is described in detail in the appropriate sections dealing with these elements.
Metabolic control elements
In a screen of over 20,000 independent insertion mutants for host factors that influence IS903 transposition the Derbyshire lab isolated more than 100 mutants that increased or decreased transposition and also altered its timing during colony growth[104]. These included independent mutations in a gene required for fermentative metabolism during anaerobic growth resulting in “early” transposition during colony growth and was suppressed by addition of fumarate, and other mutations in genes associated with DNA metabolism, intermediary metabolism, transport, cellular redox, protein folding and proteolysis. Other mutations were isolated in pur genes involved in purine biosynthesis. Further analysis suggested that this phenotype was due to a requirement for GTP in IS903 transposition[105]. It should be noted that some of these mutants also affected transposition of IS10 and of Tn552.
Hfq
Finally, the RNA chaperone Hfq has also been implicated in the regulation of Tn10 transposition by promoting RNAout interaction with transposase mRNA[106][107][108]. There is also some suggestion that SmAP1, the archaeal Hfq homolog, may also play a role in regulation of transposition of several different archaeal IS [109] but this remains to be tested directly.
Over-production inhibition
Certain transposons appear to be subject to a mode of regulation known as over-expression inhibition. This was first observed with the eukaryotic transposons Tc1/mariner Lampe[110][111][112] where increasing the concentration of transposase results in a reduction in the level of transposition. It was subsequently observed with the sleeping beauty transposon[113][114]. It also occurs in vivo in mice[115][116].
The biological rational for this is that “infection” of a naïve cell by the transposon results in a burst of transposition which is then attenuated by overproduction inhibition. This is then followed by gradual decay of the transposon.
The Chalmers lab[117] has provided an interesting and compelling explanation of this effect. Using the mariner family transposon Hsmar1 they present convincing data implying that overproduction inhibition occurs during transpososome assembly and is due to a combination of the multimeric state of the transposase coupled with competition for transposase binding sites at the Hsmar1 ends[118][119]. The model (assembly-site-occlusion model) is based on the presence of transposase multimers (dimers) to the exclusion of monomers – in other words, end-binding required a dimeric transposase. At low transposase/transposon ratios, one dimer can bind both transposon ends resulting in the ordered assembly of the transpososome. An increase in the transposase dimer/transposon ratio results in binding of dimers to both transposon ends, preventing transpososome assembly. The model not only explains the in vivo transposase dose-response for Hsmar1 but also for the related Sleeping Beauty (SB) and piggyBac (PB) transposons. As yet, no information is at present available concerning the relevance of this mode of regulation to prokaryotic transposable elements.
Bibliography
- ↑ <pubmed>2553980</pubmed>
- ↑ 2.0 2.1 <pubmed>1848178</pubmed>
- ↑ <pubmed>8107136</pubmed>
- ↑ 4.0 4.1 4.2 <pubmed>8556869</pubmed>
- ↑ 5.0 5.1 <pubmed>6281761</pubmed>
- ↑ <pubmed>6313938</pubmed>
- ↑ 7.0 7.1 <pubmed>3015876</pubmed>
- ↑ <pubmed>2416461</pubmed>
- ↑ 9.0 9.1 <pubmed>2438419</pubmed>
- ↑ <pubmed>7934941</pubmed>
- ↑ 11.0 11.1 <pubmed>8384687</pubmed>
- ↑ <pubmed>8852897</pubmed>
- ↑ <pubmed>8811194</pubmed>
- ↑ <pubmed>2543983</pubmed>
- ↑ <pubmed>1660923</pubmed>
- ↑ <pubmed>PMC1088944</pubmed>
- ↑ <pubmed>16460832</pubmed>
- ↑ <pubmed>21673094</pubmed>
- ↑ <pubmed>10762254</pubmed>
- ↑ <pubmed>15788401</pubmed>
- ↑ <pubmed>23185002</pubmed>
- ↑ <pubmed>15372017</pubmed>
- ↑ <pubmed>12029131</pubmed>
- ↑ <pubmed>1963949</pubmed>
- ↑ <pubmed>8409920</pubmed>
- ↑ <pubmed>8387998</pubmed>
- ↑ 27.0 27.1 27.2 27.3 <pubmed>2161528</pubmed>
- ↑ 28.0 28.1 <pubmed>8898394</pubmed>
- ↑ <pubmed>1101028</pubmed>
- ↑ <pubmed>378977</pubmed>
- ↑ <pubmed>9302015</pubmed>
- ↑ <pubmed>17078817</pubmed>
- ↑ <pubmed>6299577</pubmed>
- ↑ <pubmed>6291787</pubmed>
- ↑ 35.0 35.1 <pubmed>6271455</pubmed>
- ↑ 36.0 36.1 <pubmed>22195971</pubmed>
- ↑ <pubmed>15207871</pubmed>
- ↑ <pubmed>7692216</pubmed>
- ↑ <pubmed>8412678</pubmed>
- ↑ <pubmed>6296834</pubmed>
- ↑ <pubmed>2826132</pubmed>
- ↑ <pubmed>15469518</pubmed>
- ↑ <pubmed>2154486</pubmed>
- ↑ <pubmed>10677279</pubmed>
- ↑ <pubmed>11352577</pubmed>
- ↑ <pubmed>9761671</pubmed>
- ↑ <pubmed>8132525</pubmed>
- ↑ <pubmed>22032517</pubmed>
- ↑ <pubmed>22277150</pubmed>
- ↑ <pubmed>1655409</pubmed>
- ↑ <pubmed>9867814</pubmed>
- ↑ <pubmed>18680433</pubmed>
- ↑ <pubmed>12446721</pubmed>
- ↑ <pubmed>17157316</pubmed>
- ↑ <pubmed>19609359</pubmed>
- ↑ <pubmed>19229293</pubmed>
- ↑ <pubmed>26405228</pubmed>
- ↑ <pubmed>23135398</pubmed>
- ↑ <pubmed>10884228</pubmed>
- ↑ <pubmed>8805293</pubmed>
- ↑ <pubmed>2995832</pubmed>
- ↑ <pubmed>6261245</pubmed>
- ↑ <pubmed>7744253</pubmed>
- ↑ 64.0 64.1 <pubmed>9630232</pubmed>
- ↑ <pubmed>7556079</pubmed>
- ↑ <pubmed>2844529</pubmed>
- ↑ <pubmed>11447129</pubmed>
- ↑ <pubmed>14530435</pubmed>
- ↑ <pubmed>14992723</pubmed>
- ↑ 70.0 70.1 <pubmed>19696075</pubmed>
- ↑ <pubmed>19042975</pubmed>
- ↑ 72.0 72.1 72.2 72.3 <pubmed>16166383</pubmed>
- ↑ 73.0 73.1 <pubmed>17501923</pubmed>
- ↑ <pubmed>7504907</pubmed>
- ↑ <pubmed>10583504</pubmed>
- ↑ 76.0 76.1 <pubmed>9077464</pubmed>
- ↑ 77.0 77.1 <pubmed>9348666</pubmed>
- ↑ 78.0 78.1 <pubmed>9755182</pubmed>
- ↑ <pubmed>8022280</pubmed>
- ↑ <pubmed>8631314</pubmed>
- ↑ <pubmed>7557391</pubmed>
- ↑ <pubmed>9203582</pubmed>
- ↑ <pubmed>8617219</pubmed>
- ↑ <pubmed>9299335</pubmed>
- ↑ <pubmed>7932694</pubmed>
- ↑ <pubmed>9729608</pubmed>
- ↑ 87.0 87.1 <pubmed>1648004</pubmed>
- ↑ 88.0 88.1 <pubmed>1655708</pubmed>
- ↑ 89.0 89.1 <pubmed>1657870</pubmed>
- ↑ <pubmed>1328165</pubmed>
- ↑ 91.0 91.1 91.2 <pubmed>3025455</pubmed>
- ↑ <pubmed>10844241</pubmed>
- ↑ <pubmed>10715006</pubmed>
- ↑ <pubmed>6267432</pubmed>
- ↑ <pubmed>6288966</pubmed>
- ↑ <pubmed>3030303</pubmed>
- ↑ <pubmed>6290084</pubmed>
- ↑ <pubmed>6265907</pubmed>
- ↑ <pubmed>7592347</pubmed>
- ↑ <pubmed>8930913</pubmed>
- ↑ 101.0 101.1 <pubmed>3000598</pubmed>
- ↑ <pubmed>2451025</pubmed>
- ↑ <pubmed>10207011</pubmed>
- ↑ <pubmed>16135227</pubmed>
- ↑ <pubmed>15968071</pubmed>
- ↑ <pubmed>23510801</pubmed>
- ↑ <pubmed>25649688</pubmed>
- ↑ <pubmed>25579599</pubmed>
- ↑ <pubmed>36912639</pubmed>
- ↑ <pubmed>9584095</pubmed>
- ↑ <pubmed>8882498</pubmed>
- ↑ <pubmed>9154003</pubmed>
- ↑ <pubmed>12842434</pubmed>
- ↑ <pubmed>14759813</pubmed>
- ↑ <pubmed>14961361</pubmed>
- ↑ <pubmed>14529839</pubmed>
- ↑ <pubmed>23795293</pubmed>
- ↑ <pubmed>24812590</pubmed>
- ↑ <pubmed>26104691</pubmed>