Difference between revisions of "General Information/Fuzzy Borders"
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
==Fuzzy Borders== | ==Fuzzy Borders== | ||
{| | {| | ||
| − | |'''<big>W</big>'''ith our increasing knowledge of the diversity of the mobilome, the distinction between IS and other TE is becoming increasing unclear. The major feature used to distinguish IS from transposons was that the former [[:Image:1.13.1.png|(Fig. | + | |'''<big>W</big>'''ith our increasing knowledge of the diversity of the mobilome, the distinction between IS and other TE is becoming increasing unclear. The major feature used to distinguish IS from transposons was that the former [[:Image:1.13.1.png|(Fig.8.1)]] lack phenotypically detectable passenger genes (genes not involved in the transposition process) while the latter include one or more such genes (for antibiotic resistance, virulence and pathogenicity functions or genes permitting the use of unusual compounds). This is no longer the case [[:Image:1.13.1.png|(Fig.8.1)]]. |
| − | As shown in [[:Image:1.13.1.png|Fig. | + | As shown in [[:Image:1.13.1.png|Fig.8.1]], examples have now been identified in which passenger genes are located within an IS, called tIS (ISs and relatives with passenger genes)<ref><nowiki><pubmed>19286454</pubmed></nowiki></ref> or in which TE with typical transposon structures are devoid of transposition proteins MITEs ('''M'''iniature '''I'''nverted repeat '''T'''ransposable '''E'''lements) and MICs ('''M'''obile '''I'''nsertion '''C'''assette))<ref><nowiki><pubmed>15228527</pubmed></nowiki></ref><ref><nowiki><pubmed>10320586</pubmed></nowiki></ref><ref><nowiki><pubmed>11814653</pubmed></nowiki></ref><ref><nowiki><pubmed>12520302</pubmed></nowiki></ref>. The (hypothetical) relationship between these different autonomous (with transposase) and non-autonomous IS derivatives is indicated in [[:Image:1.13.1.png|Fig.8.1]] <ref><nowiki><pubmed>24499397</pubmed></nowiki></ref><ref><nowiki><pubmed>26104715</pubmed></nowiki></ref>.<br /><br /><br /><br /><br /><br /><br /><br /><br /><br /> |
| − | |[[Image:1.13.1.png|thumb|480x480px|'''Fig. | + | |[[Image:1.13.1.png|thumb|480x480px|'''Fig.8.1.''' Relationship between IS, tIS, and MITES. The (hypothetical) relationship between different IS derivatives (shown as pink boxes). Horizontal arrows indicate open reading frames encoding the Tpase (purple), passenger genes (red). The terminal inverted repeats are shown as dark blue triangles. Examples of IS families which include such derivatives are: [[IS Families/IS4 and related families#IS231|IS''4'' grp IS''231'']], [[IS Families/IS1595 family|IS''1595'']], and [[IS Families/IS66 family|IS''66'']].|alt=|none]] |
|} | |} | ||
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 12:46, 23 June 2020
Fuzzy Borders
| With our increasing knowledge of the diversity of the mobilome, the distinction between IS and other TE is becoming increasing unclear. The major feature used to distinguish IS from transposons was that the former (Fig.8.1) lack phenotypically detectable passenger genes (genes not involved in the transposition process) while the latter include one or more such genes (for antibiotic resistance, virulence and pathogenicity functions or genes permitting the use of unusual compounds). This is no longer the case (Fig.8.1).
As shown in Fig.8.1, examples have now been identified in which passenger genes are located within an IS, called tIS (ISs and relatives with passenger genes)[1] or in which TE with typical transposon structures are devoid of transposition proteins MITEs (Miniature Inverted repeat Transposable Elements) and MICs (Mobile Insertion Cassette))[2][3][4][5]. The (hypothetical) relationship between these different autonomous (with transposase) and non-autonomous IS derivatives is indicated in Fig.8.1 [6][7]. |
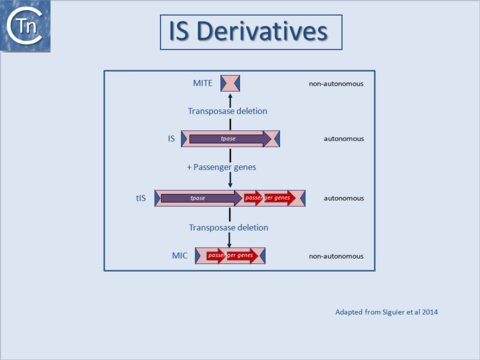 Fig.8.1. Relationship between IS, tIS, and MITES. The (hypothetical) relationship between different IS derivatives (shown as pink boxes). Horizontal arrows indicate open reading frames encoding the Tpase (purple), passenger genes (red). The terminal inverted repeats are shown as dark blue triangles. Examples of IS families which include such derivatives are: IS4 grp IS231, IS1595, and IS66. |