General Information/IS and Gene Expression
Another important aspect of IS impact on their bacterial hosts is their ability to modulate gene expression. In addition to acting as vectors for gene transmission from one replicon to another in the form composite transposons (two IS flanking any gene; Fig.1.2.3) and tIS (Fig.1.13.1) and their ability to interrupt genes, it has been known for some time[1][2] that IS can also activate gene expression. This capacity has recently received much attention due to the increase in resistance to various antibacterials[3][4][5], a worrying public health threat[6][7].
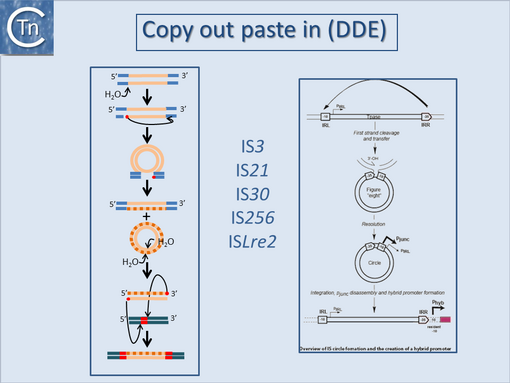
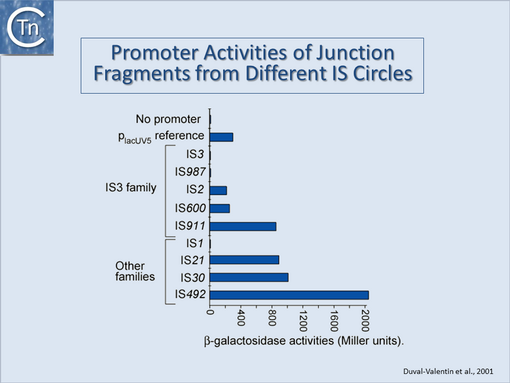
They can accomplish this in two ways: either by providing internal promoters whose transcripts escape into neighboring DNA[2][8][9][10] or by hybrid promoter formation. Many IS carry -35 promoter components oriented towards the flanking DNA (Fig.1.24.1). In a number of cases this plays an important part in their transposition since a significant number of IS transpose using an excised transposon circle (Fig.1.24.1) with abutted left and right ends. For these IS, the other end carries a -10 element oriented inwards towards the Tpase gene. Together with the -35, this generates a strong promoter on formation of the circle junction to drive Tpase expression required for catalysis of integration (Fig.1.24.2) [11][12][13][14]. Thus if integration occurs next to a resident -10 sequence, the IS -35 sequence can contribute to a hybrid promoter to drive expression of neighboring genes [see [15]]. At present this phenomenon had been reported to occur with over 30 different IS in more than 17 bacterial species[16][17] (Table IS and Gene Expression below). Indeed, specific vector plasmids have been designed to identify activating insertions (e.g. [18]).
IS activity can affect efflux mechanisms resulting in increased resistance: IS1 or IS10 insertion can up-regulate the AcrAB-TolC pump in Salmonella enterica[19]; IS1 or IS2 insertion upstream of AcrEF[20][21] and IS186 insertional inactivation of the AcrAB repressor, AcrR, in Escherichia coli [20], all lead to increased resistance to fluoroquinolones. Insertional inactivation of specific porins can also play a significant role[22].
IS and Gene Expression
| Table. IS and gene expression. | ||||||||
| IS family | IS name | Mechanism | Gene(s) affected | Organism | Reference | Clinical/Experimental | ||
|---|---|---|---|---|---|---|---|---|
| IS1 | IS1 | Cointegrate | EmR | Escherichia coli | [23] | C? | ||
| Copy-paste circles | blaTEM-1 | [15] | E | |||||
| — | acrEF pump | Salmonella enterica | [19] | E | ||||
| IS3 | IS2 | Copy-paste circles | gal | Escherichia coli | [24][25] | E | ||
| gal | [23][2] | E | ||||||
| argE | [26][20] | E | ||||||
| EmR | C? | |||||||
| blaampC | E | |||||||
| acrEF pump | E | |||||||
| IS3 | Copy-paste circles | argE | Escherichia coli | [2] | E | |||
| argE | [8] | E | ||||||
| citT | [27] | E | ||||||
| IS981 | Copy-paste circles | ldhB | Lactococcus lactis | [28] | E | |||
| IS6110 | Copy-paste circles | Rv2280 and PE-PGRS gene, Rv1468c | Mycobacterium tuberculosis | [29] | E | |||
| ISKpn8 | Copy-paste circles | blaKCP-2 | Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Enterobacter aerogenes, and Klebsiella oxytoca | [30] | C | |||
| IS4 | IS10 | Cut-paste (hairpin) | his | Salmonella typhimurium | [31] | E | ||
| Cut-paste | — | Escherichia coli | [32] | E | ||||
| Cut-paste | acrEF pump | Salmonella enterica | [19] | E | ||||
| IS50 | Cut-paste (hairpin) | aph3’II | Escherichia coli | [33] | E | |||
| IS1999 | — | blaVEB-1 | Pseudomonas aeruginosa | [4] | C | |||
| blaVEB-1/blaOXA-48 | Escherichia coli | [3] | C | |||||
| ISPa12 | — | blaPER-1 | Salmonella enterica, Pseudomonas aeruginosa, Providencia stuartii, Acinetobacter baumannii | [34] | C | |||
| ISAba1 | — | blaampC | Acinetobacter baumannii | [35][36][37] | C | |||
| blaOXA-51/blaOXA-23 | [38] | C | ||||||
| IS5 | IS5 | — | EmR | Escherichia coli | [23] | C? | ||
| ISFtu2 | general | Francisella tularensis | [39] | Natural isolate | ||||
| ISVa1 | (iron uptake) | Vibrio anguilarum | [40] | Natural isolate | ||||
| IS1168 (IS1186) | nimA, nimB | Bacteroides sp. | [41] | C | ||||
| IS1186 | cfiA | Bacteroides fragilis | [42][43] | C | ||||
| IS402 | bla | Pseudomonas cepacia | [44] | E | ||||
| IS6 | IS257 | Cointegrate | dfrA | Staphylococcus aureus | [45] | C | ||
| IS257 | Cointegrate | tet | Staphylococcus aureus | [46] | ||||
| IS1008 | — | blaOXA-58 | Acinetobacter baumannii | [47] | ||||
| IS26 | — | aphA7, blaS2A | Klebsiella pneumoniae | [48] | ||||
| blaSHV-2a | Pseudomonas aeruginosa | [49] | ||||||
| IS140 | — | aac(3)-III and -IV | — | [50] | ||||
| IS21 | ISBf1 | Copy-paste circle | cepA | Bacteroides fragilis | [51] | C | ||
| ISKpn7 | blaKPC | Klebsiellea pneumonia | [52] | |||||
| IS30 | IS30 | Copy-paste circle | galK | Escherichia coli | [53] | E | ||
| IS18 | aac(6’)-Ij | Acinetobacter sp. | [54] | C | ||||
| blaOXA257 | [55] | |||||||
| IS4351 | ermF | Bacteroides fragilis | [56] | C | ||||
| cfiA | [5] | |||||||
| IS1086 | cnrCBAT (ZnR) | Cupriavidus metallidurans | E | |||||
| IS256 | IS256 | Copy-paste circles | mecA | Staphylococcus sciuri | [57][58] | — | ||
| llm | Staphylococcus aureus | |||||||
| IS1490 | — | Burkholderia cepacia | [59] | — | ||||
| IS406 | — | Burkholderia cepacia | [44] | E | ||||
| IS481 | IS481 | Copy-paste circles | katA | Bordetella pertussis | [60] | — | ||
| ISRme5 | cnrCBAT (Zn R) | Cupriavidus metallidurans | [61] | E | ||||
| IS630 | ISFtu1 | Tc-like | general | Francisella tularensis | [39] | Natural isolate | ||
| IS982 | IS1187 | — | cfiA | Bacteroides fragilis | [42] | E+C | ||
| [5][62][63] | C | |||||||
| IS982 | citQRP | Lactococcus lactis | [64] | — | ||||
| IS1380 | ISBf12 | — | cfiA | Bacteroides fragilis | [5] | C | ||
| IS612 | [63] | |||||||
| IS613 | [63] | |||||||
| IS614 | [5][63][65] | |||||||
| IS1188 | [62] | |||||||
| IS942 | [62] | |||||||
| ISEcp1 | blaCTX-M-15 | Enterobacteriaceae | [66] | |||||
| blaCTX-M-17 | Klebsiella pneumoniae | [67] | ||||||
| blaCTX-M-19 | Klebsiella pneumoniae | [34] | ||||||
| blaCTX-M | Kluyvera ascorbata | [68] | ||||||
| rmtC | Escherichia coli | [69] | ||||||
| IS1187 | cfiA | Bacteroides fragilis | [62] | |||||
| ISL3 | ISSg1 | Copy-paste circles | sspB (surface antigen) | Streptococcus gordonii | [70] | — | ||
| IS1411 | pheBA | Pseudomonas putida | [71] | |||||
| ISAs1 | IS1548 | — | lmb (lamelin binding) | Streptococcus agalactiae | [72] | — | ||
| nd | — | Acinetobacter baumannii | [35] | C | ||||
| ISNYC | IS403 | — | bla | Burkholderia cepacia | [44] | E | ||
| IS404 | ||||||||
| IS405 | ||||||||
Bibliography
- ↑ <pubmed>1101028</pubmed>
- ↑ 2.0 2.1 2.2 2.3 Glansdorff N, Charlier D, Zafarullah M . Activation of gene expression by IS2 and IS3. - Cold Spring Harb Symp Quant Biol: 1981, 45 Pt 1;153-6 [PubMed:6271458] [DOI] </nowiki>
- ↑ 3.0 3.1 </nowiki>
- ↑ 4.0 4.1 Aubert D, Naas T, Nordmann P . IS1999 increases expression of the extended-spectrum beta-lactamase VEB-1 in Pseudomonas aeruginosa. - J Bacteriol: 2003 Sep, 185(17);5314-9 [PubMed:12923109] [DOI] </nowiki>
- ↑ 5.0 5.1 5.2 5.3 5.4 </nowiki>
- ↑ <pubmed>23887414</pubmed>
- ↑ <pubmed>23887415</pubmed>
- ↑ 8.0 8.1 </nowiki>
- ↑ <pubmed>6260746</pubmed>
- ↑ <pubmed>6311437</pubmed>
- ↑ <pubmed>26350305</pubmed>
- ↑ <pubmed>9214651</pubmed>
- ↑ <pubmed>10438765</pubmed>
- ↑ <pubmed>11598022</pubmed>
- ↑ 15.0 15.1 </nowiki>
- ↑ <pubmed>17223624</pubmed>
- ↑ <pubmed>24499397</pubmed>
- ↑ <pubmed>8863735</pubmed>
- ↑ 19.0 19.1 19.2 Olliver A, Vallé M, Chaslus-Dancla E, Cloeckaert A . Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar typhimurium DT204 acrB mutants selected with fluoroquinolones. - Antimicrob Agents Chemother: 2005 Jan, 49(1);289-301 [PubMed:15616308] [DOI] </nowiki>
- ↑ 20.0 20.1 20.2 </nowiki>
- ↑ <pubmed>11274125</pubmed>
- ↑ <pubmed>15212803</pubmed>
- ↑ 23.0 23.1 23.2 </nowiki>
- ↑ <pubmed>4610339</pubmed>
- ↑ <pubmed>339095</pubmed>
- ↑ <pubmed>6187472</pubmed>
- ↑ <pubmed>22992527</pubmed>
- ↑ <pubmed>12867459</pubmed>
- ↑ <pubmed>15130120</pubmed>
- ↑ <pubmed>24433026</pubmed>
- ↑ <pubmed>6289329</pubmed>
- ↑ <pubmed>6311437</pubmed>
- ↑ <pubmed>6260374</pubmed>
- ↑ 34.0 34.1 </nowiki>
- ↑ 35.0 35.1 </nowiki>
- ↑ <pubmed>14742218</pubmed>
- ↑ <pubmed>16441449</pubmed>
- ↑ <pubmed>16630258</pubmed>
- ↑ 39.0 39.1 </nowiki>
- ↑ <pubmed>7568465</pubmed>
- ↑ <pubmed>8067736</pubmed>
- ↑ 42.0 42.1 </nowiki>
- ↑ <pubmed>7545155</pubmed>
- ↑ 44.0 44.1 44.2 </nowiki>
- ↑ <pubmed>7840551</pubmed>
- ↑ <pubmed>10852863</pubmed>
- ↑ <pubmed>18443121</pubmed>
- ↑ <pubmed>2160941</pubmed>
- ↑ <pubmed>10223953</pubmed>
- ↑ <pubmed>6318050</pubmed>
- ↑ <pubmed>7517394</pubmed>
- ↑ <pubmed>18227185</pubmed>
- ↑ <pubmed>3039299</pubmed>
- ↑ <pubmed>9756793</pubmed>
- ↑ <pubmed>23014718</pubmed>
- ↑ <pubmed>3038844</pubmed>
- ↑ <pubmed>9371438</pubmed>
- ↑ <pubmed>12511511</pubmed>
- ↑ <pubmed>9098071</pubmed>
- ↑ <pubmed>7830550</pubmed>
- ↑ <pubmed>27047473</pubmed>
- ↑ 62.0 62.1 62.2 62.3 Podglajen I, Breuil J, Rohaut A, Monsempes C, Collatz E . Multiple mobile promoter regions for the rare carbapenem resistance gene of Bacteroides fragilis. - J Bacteriol: 2001 Jun, 183(11);3531-5 [PubMed:11344163] [DOI] </nowiki>
- ↑ 63.0 63.1 63.2 63.3 </nowiki>
- ↑ <pubmed>8602160</pubmed>
- ↑ <pubmed>19744834</pubmed>
- ↑ <pubmed>11470367</pubmed>
- ↑ <pubmed>12435670</pubmed>
- ↑ <pubmed>16569841</pubmed>
- ↑ <pubmed>16940134</pubmed>
- ↑ <pubmed>9202480</pubmed>
- ↑ <pubmed>9765560</pubmed>
- ↑ <pubmed>20520730</pubmed>