Glossary
A
B
C
- Class I Elements: (see also)
- Class II Elements: (see also)
- Cre Recombinase: The Cre Recombinase is a DNA breaking and rejoining enzyme (from tyrosine recombinase family) derived from the P1 bacteriophage. It is widely used for conditional mutagenesis of transgenes and insert DNA cassettes into eukaryotic chromosomes. (see also).
- Co-integrate:
- Copy-and-paste transposition model:
- Copy-out–Paste-in transposition model:
- Cut-and-paste transposition model:
D
- Direct Repeats (DRs):
- DDE Domain:
- Donor Primed Replicative Transposition (DPRT):
E
F
G
- Group II introns: The group II introns are mobile retroelements that use the combined activities of an autocatalytic RNA and an intron-encoded reverse transcriptase (RT) to propagate efficiently within genomes. They are intimately related with the evolution of eukaryotes, being ancestrally related to nuclear spliceosomal introns, retrotransposons and telomerase (see also).
- Genome decay:
- Genome Rearrangements:
H
- Horizontal gene transfer (or Lateral gene transfer):
- Homologous recombination:
I
- Inverted Repeats (IR):
- Integron: Integrons one class of Mobile Genetic Elements (MGEs) are specific genetic structure composed by genes (named as integron cassettes) that generally allow bacteria to adapt and rapidly evolve through the acquisition, stockpiling, excision, and reordering of open reading frames found the integron cassettes. The integron mobilization is mediated by site-specific recombination reactions by the integrase (see also).
- Invertases:
J
K
L
M
- Mobilome:
- Methylase (Methylation):
N
O
P
- Plasmidome:
- plasmid F:
- Pseudogenisation:
- Peel-and-paste (Single-strand) transposition model:
- Phage tyrosine integrase:
- Phage serine integrase:
Q
R
- Resolution site:
- Resolvase:
- Rolling-circle transposition model:
S
- Synaptic complex:
- Serine recombinase: The serine recombinases are a family of DNA breaking and rejoining enzymes. Unlike homologous recombination, the serine recombinases promote rearrangements of DNA by breaking and rejoining strands at precisely defined sequence positions (see also).
- Serine Resolvases: The serine resolvases and the closely related invertases, are a group of site-specific recombinases that, in their native contexts, resolve large fused replicons into smaller separated ones
- Single-strand transposition:
- Site-specific recombination:
T
- Target specificity:
- Transposase:
- Transposome:
- Target Primed Replicative Transposition (TPRT):
- Tyrosine recombinase: Tyrosine site-specific recombinases (YRs) are a family of DNA breaking and rejoining enzymes which use the active site tyrosine nucleophile for DNA strand breakage (see also).
U
V
- V(D)J Recombination:
W
X
- Xer Site-Specific Recombination:
Y
Z
See also
The logic behind the TEs nomenclature and naming attribution
Why should researchers contemplate naming a newly identified TE?
Mainly to name it, so the discoverer and other researchers can specifically refer to it [1].
- Prokaryotic TE nomenclature and naming attribution
Transposable elements were discovered by Barbara McClintock during experiments conducted in 1944 on maize. However, her discovery was met with less than enthusiastic reception by the genetic community [2] (for a full detailed history, please see Evelyn Fox Keller's acclaimed biography: A Feeling for the Organism, 10th Aniversary Edition: The Life and Work of Barbara McClintock). Only some decades later, the her discovery was brought back to life after the Szybalski group in the early 1970s discovered the bacterial insertion sequences [2][3], and the picture of transposable elements started to change.
A committee assembled during the meeting on DNA Insertions at Cold Spring Harbor in 1976 proposed a set of rules to be used for the nomenclature of Transposable Elements [4]. The first attempt to create an concise nomenclature system for Transposable Elements started at Stanford University by Campbell and colleagues in 1979 [5]. They classified the prokaryotes elements as simple IS elements to more complex Tn transposons, and self-replicating episomes. In addition, definitions and nomenclature rules for these three classes of prokaryotic TEs were specified [5] (Table 1).
| Classes | Definition | Naming rule |
|---|---|---|
| IS elements | IS elements contain no known genes unrelated to insertion functions, and are shorter than 2kb. | IS1, IS2, IS3, IS4, IS5, etc, with the numbers italicized |
| Tn elements | Tn are more complex, often containing two copies of an IS element. They are generally larger than 2kb, and contain additional genes unrelated to insertion functions. | Tn1, Tn2, Tn3, etc, with the numbers italicized |
| Episomes | Episomes are complex, self-replicating elements, often containing IS and Tn elements. Examples include the phage λ and plasmid F of E. coli. | - |
The allocation of numbers and database administration was carried out by the late Dr Esther Lederberg from Stanford University Medical School, CA, USA [4]. Lists for the registry of Tn number allocations were subsequently published [6]. The allocation of Tn numbers stopped with the retirement of Dr Lederberg and gradually a variety of rules were adopted for naming newly discovered transposons [4]. At the same time new types of transposable elements, such as the mobilizable and conjugative transposons, were being discovered [4]. Additionally, interactions between different elements including transposition and/or recombination events led to novel chimeric transposons. These exacerbated the nomenclature problem [4]. Subsequent nomenclature systems have become complicated, with different systems being adopted for related elements by different research groups [4]. Therefore, Robert and colleagues in 2008 proposed a new version of the early nomenclature system, but not including non-autonomous elements (such as integron cassettes and MITEs) [4] (see below).
| Type of transposable element a | Definition |
|---|---|
| Composite transposons | Flanked by IS elements. The transposase of the IS element is responsible for the catalysis of insertion and excision. |
| Unit transposons | Typical unit elements encode an enzyme involved in excision and integration (DD(35)E or tyrosine) often a site specific recombinase or resolvase and one or several accessory (e.g. resistance) genes in one genetic unit. |
| Conjugative transposons (CTns) / Integrative conjugative elements (ICEs) | The conjugative transposons (CTns), also known as integrative conjugative elements (ICEs), carry genes for excision, conjugative transfer and for integration within the new host genome. They carry a wide range of accessory genes, including antibiotic resistance |
| Mobilisable transposons (MTns) / Integrative mobilisable elements (IMEs) | The mobilizable transposons (MTns), also known as integrative mobilizable elements (IMEs), can be mobilized between bacterial cells by other “helper”elements that encode proteins involved in the formation of the conjugation pore or mating bridge. The MTns exploit these conjugation pores and generally provide their own DNA processing functions for intercellular transfer and subsequent transposition. |
| Mobile genomic islands | Some chromosomally integrated genomic islands encode tyrosine or serine site-specific recombinases that catalyze their own excision and integration but do not harbor genes involved in transfer. They carry genes encoding for a range of phenotypes. The name of a genomic island reflects the phenotype it confers, e.g. pathogenicity islands encode virulence determinants (toxins, adhesins etc). |
| Integrated or transposable prophage | An integrated or transposable prophage is a phage genome inserted as part of the linear structure of the chromosome of a bacterium which is able to excise and insert from and into the genome. |
| Integrated satellite prophage | Bacteriophage genome inserted into that of the host which requires gene products from “helper” phages to complete its replication cycle |
| Group I intron | Small post-transcriptionally splicing (splicing occurs in the pre-mRNA), endonuclease encoding element. Will home to allelic site |
| Group II intron | Small post-transcriptionally splicing (splicing occurs in the pre-mRNA), restriction endonuclease encoding element |
| IStron intein | Chimeric ribozyme consisting of a group I intron linked to an IS605 like transposase Small post-translational splicing (splicing occurs in the polypeptide), endonuclease encoding element. Will home to allelic site. |
a; not all reported elements have been shown to be mobile
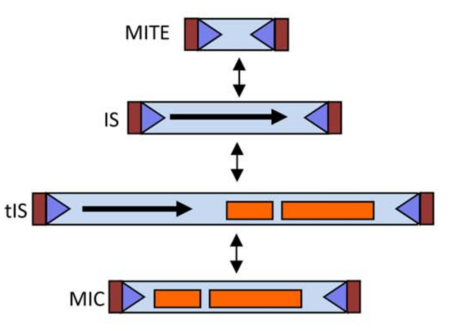
Following Roberts et al [4] classification system, the ISfinder team (Mick Chandler and Patricia Siguier) and Jacques Mahillon proposed a enhancement for this classification system, now including terms and definitions for MICs (Mobile Insertion Cassette), composed of passenger genes but no transposase, and MITES (Miniature Inverted repeat Transposable Element), which are short IS-related elements with no internal open reading frame but which, like most ISs, transposons and MICs, include IS-like extremities. It is important to note that a single IS might in principle be represented in all four forms (see the table and figure below).
| Extremities | Transposase | Passenger genes | |
|---|---|---|---|
| MITE | yes | no | no |
| IS | yes | yes | no |
| tIS | yes | yes | yes |
| MIC | yes | no | yes |
Bioinformatics approaches for prokaryotic TE identification and annotation
- Eukaryotic TE nomenclature and naming attribution
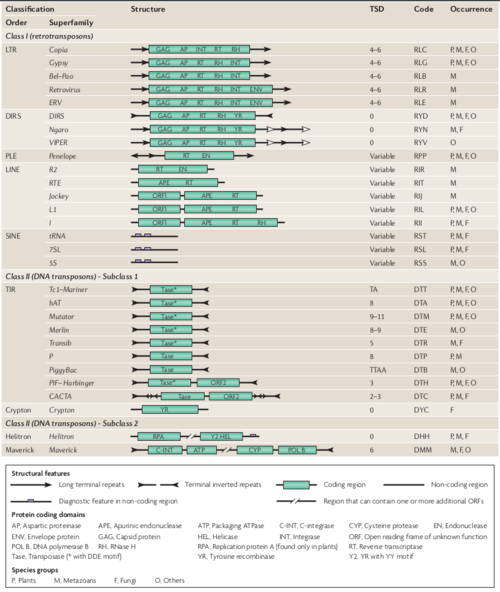
Since TEs have been found in virtually all eukaryotic species investigated so far, displaying an extreme diversity, revealed by thousands or even tens of thousands of different TE families[7], Finnegan and colleagues in 1989, proposed the first TE classification system, which distinguished two classes by their transposition intermediate: RNA (Class I or retrotransposons) or DNA (Class II or DNA transposons)[7][8]. The transposition mechanism of Class I is commonly called ‘copy-and-paste’, and that of Class II, ‘cut-and-paste’ [8]. Later, in 2007, Wicker and colleagues proposed a common TE classification system that can be easily handled by non-specialists during genome and TEs annotation procedures [7]. This system provided a consensus between the various conflicting classification and naming systems that are currently in use[7]. A key component of this system is a naming convention: a three-letter code with each letter respectively denoting class, order and superfamily; the family (or subfamily) name; the sequence (database accession number) on which the element was found; and the ‘running number’, which defines the individual insertion in the accession. The unified system is also intended to facilitate comparative and evolutionary studies on TEs from different species. (see figure below)[7] .
Nowadays, the Wicker TE classification system is well-recognized by the scientific community, and are currently in use by the TE annotation pipelines.
Bioinformatics approaches for eukaryotic TE identification and annotation