Difference between revisions of "IS Families/IS5 and related IS1182 families"
| Line 1: | Line 1: | ||
====IS''5'' family==== | ====IS''5'' family==== | ||
=====Original Identification===== | =====Original Identification===== | ||
| − | IS''5'' was originally isolated as an insertion into the immunity region of [[wikipedia:Lambda_phage|bacteriophage lambda]] and subsequently found as a cause of mutation in a number of ''[[wikipedia:Escherichia_coli|E. coli]]'' genes<ref><nowiki><pubmed>4432374</pubmed></nowiki></ref><ref><nowiki><pubmed>353507</pubmed></nowiki></ref><ref><nowiki><pubmed>84614</pubmed></nowiki></ref><ref><nowiki><pubmed>641012</pubmed></nowiki></ref>. Together with IS''1'', it was also identified as an activator (by insertion) of expression of the usually cryptic [[wikipedia:Beta-glucosidase|beta-glucosidase]] gene of ''[[wikipedia:Escherichia_coli|E. coli]]''<ref><nowiki><pubmed>6270569</pubmed></nowiki></ref><ref><nowiki><pubmed>3034860</pubmed></nowiki></ref><ref name=":0"><pubmed>1311089</pubmed> | + | IS''5'' was originally isolated as an insertion into the immunity region of [[wikipedia:Lambda_phage|bacteriophage lambda]] and subsequently found as a cause of mutation in a number of ''[[wikipedia:Escherichia_coli|E. coli]]'' genes<ref><nowiki><pubmed>4432374</pubmed></nowiki></ref><ref><nowiki><pubmed>353507</pubmed></nowiki></ref><ref><nowiki><pubmed>84614</pubmed></nowiki></ref><ref><nowiki><pubmed>641012</pubmed></nowiki></ref>. Together with [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1R IS''1''], it was also identified as an activator (by insertion) of expression of the usually cryptic [[wikipedia:Beta-glucosidase|beta-glucosidase]] gene of ''[[wikipedia:Escherichia_coli|E. coli]]''<ref><nowiki><pubmed>6270569</pubmed></nowiki></ref><ref><nowiki><pubmed>3034860</pubmed></nowiki></ref><ref name=":0"><pubmed>1311089</pubmed> |
| + | |||
| + | </nowiki></ref><ref><nowiki><pubmed>2846278</pubmed></nowiki></ref><ref><nowiki><pubmed>7781607</pubmed></nowiki></ref><ref><nowiki><pubmed>8710516</pubmed></nowiki></ref>. | ||
=====Presence in Compound Transposons===== | =====Presence in Compound Transposons===== | ||
| − | Several members are associated with compound transposons. These include IS''903'' and IS''602'', which form part of the [[wikipedia:Kanamycin_A|kanamycin]] resistance transposons Tn''903''<ref><nowiki><pubmed>6261245</pubmed></nowiki></ref>, and Tn''602''<ref><nowiki><pubmed>2819910</pubmed></nowiki></ref> respectively, and IS''Va1''/IS''Va2'' which form part of a transposon carrying iron transport genes<ref><nowiki><pubmed>7568465</pubmed></nowiki></ref>. | + | Several members are associated with compound transposons. These include [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS602 IS''602''], which form part of the [[wikipedia:Kanamycin_A|kanamycin]] resistance transposons [http://tncentral.ncc.unesp.br/cgi-bin/tn_report.pl?id=Tn903-V00359.1 Tn''903'']<ref><nowiki><pubmed>6261245</pubmed></nowiki></ref>, and Tn''602''<ref><nowiki><pubmed>2819910</pubmed></nowiki></ref> respectively, and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISVa1 IS''Va1''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISVa2 IS''Va2''] which form part of a transposon carrying iron transport genes<ref><nowiki><pubmed>7568465</pubmed></nowiki></ref>. |
=====Distribution===== | =====Distribution===== | ||
The IS''5'' family, like the IS''4'' family, is also a relatively heterogeneous group which now requires reanalysis. It also includes sequences from both eubacteria and the archaea. | The IS''5'' family, like the IS''4'' family, is also a relatively heterogeneous group which now requires reanalysis. It also includes sequences from both eubacteria and the archaea. | ||
| − | There are now a large number of identified members of the IS''5'' family (>550 members) and of a closely related IS''1182'' family (>150 members) which have allowed a more detailed analysis and a separation into various subgroups and families. The IS''5'' family is partitioned into 6 subgroups: IS''5'', IS''903'', IS''L2'', IS''H1'', IS''1031'' and IS''427''<ref name=":1"><pubmed>9729608</pubmed> | + | There are now a large number of identified members of the IS''5'' family (>550 members) and of a closely related [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1182 IS''1182''] family (>150 members) which have allowed a more detailed analysis and a separation into various subgroups and families. The IS''5'' family is partitioned into 6 subgroups: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427'']<ref name=":1"><pubmed>9729608</pubmed> |
| − | [[Image:Fig. IS5.1.png|thumb|center| | + | |
| − | [[Image:Fig. IS5.2.png|thumb|center| | + | </nowiki></ref> ([[General Information/What Is an IS?#Characteristics%20of%20insertion%20sequence%20families|Table Characteristics of IS families]]; [[:File:Fig. IS5.1.png|Fig.5.1]]). Some of these may prove to be emerging families. Members of the IS''5'' subgroup appear to be composed of two groups with different lengths: one of 1060-1300 bp and a second of 1460-1610 ([[:File:Fig. IS5.2.png|Fig.5.2 A]]). |
| + | [[Image:Fig. IS5.1.png|thumb|center|680x680px|'''Fig. IS5.1.''' Correspondence between the IS '''IRs''' and different IS''5'' family subgroups.|alt=]] | ||
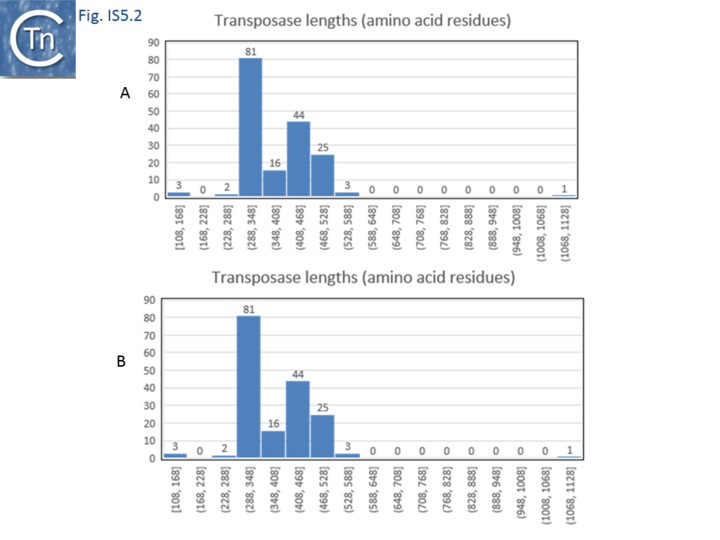
| + | [[Image:Fig. IS5.2.png|thumb|center|720x720px|'''Fig. IS5.2.''' IS''5'' family IS''5'' subgroups '''A)''' distribution of IS length (base pairs); '''B)''' distribution of the length of transposase (amino acid residues).|alt=]] | ||
=====Diversity===== | =====Diversity===== | ||
| − | The transposases of these are also of different lengths ([[:File:Fig. IS5.2.png|Fig.5.2 B]]) and transposase length is correlated with that of the IS. The lengths of the IS''1013'' subgroup are between ~900 and ~1200 bp with the majority between 103 and 1090 bp ([[:File:Fig. IS5.3.png|Fig.5.3]]), those of the IS''427'' group are between 800 and 1070 bp in length with most having lengths in the range of 810 900 bp ([[:File:Fig. IS5.3.png|Fig.5.3]]). Members of the IS''903'' subgroup are generally about 1030-1090 bp long ([[:File:Fig. IS5.4.png|Fig.5.4 A]] ), those of the IS''H1'' subgroup are about 850 - 1200 bp long (note that this subgroup includes a number of '''M'''iniature '''I'''nverted repeat '''T'''ransposable '''E'''lements (MITES) ([[:File:Fig. IS5.4.png|Fig.5.4 B]]) and IS''L2'' members are 820 to 1260 bp long with a majority of about 820-970 bp ([[:File:Fig. IS5.4.png|Fig.5.4 C]]). There are a large number of additional IS''5'' family members whose attribution to subgroups has yet to be established. | + | The transposases of these are also of different lengths ([[:File:Fig. IS5.2.png|Fig.5.2 B]]) and transposase length is correlated with that of the IS. The lengths of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1013 IS''1013''] subgroup are between ~900 and ~1200 bp with the majority between 103 and 1090 bp ([[:File:Fig. IS5.3.png|Fig.5.3]]), those of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''] group are between 800 and 1070 bp in length with most having lengths in the range of 810 900 bp ([[:File:Fig. IS5.3.png|Fig.5.3]]). Members of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] subgroup are generally about 1030-1090 bp long ([[:File:Fig. IS5.4.png|Fig.5.4 A]] ), those of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''] subgroup are about 850 - 1200 bp long (note that this subgroup includes a number of '''M'''iniature '''I'''nverted repeat '''T'''ransposable '''E'''lements (MITES) ([[:File:Fig. IS5.4.png|Fig.5.4 B]]) and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''] members are 820 to 1260 bp long with a majority of about 820-970 bp ([[:File:Fig. IS5.4.png|Fig.5.4 C]]). There are a large number of additional IS''5'' family members whose attribution to subgroups has yet to be established. |
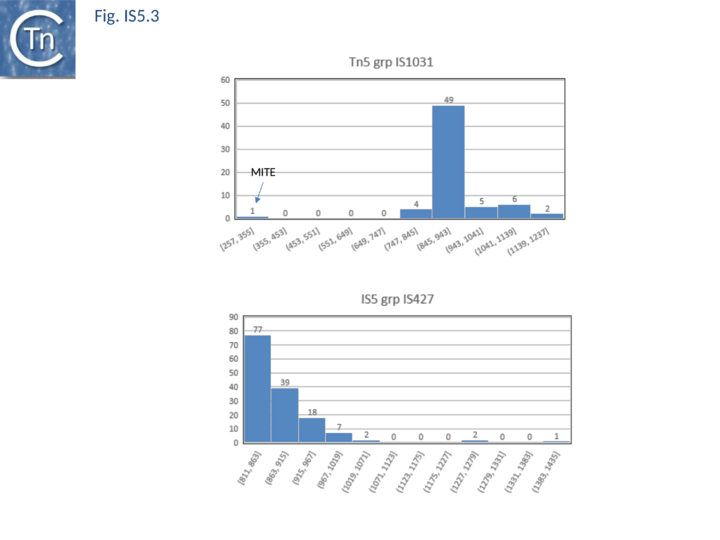
| − | [[Image:Fig. IS5.3.png|thumb|center| | + | [[Image:Fig. IS5.3.png|thumb|center|720x720px|'''Fig. IS5.3.''' IS''5'' family [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''] subgroups. '''Top''': distribution of IS length (base pairs) IS''1031''; '''Bottom''': distribution of IS length (base pairs) [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427'']. The number of examples used in the sample is shown above each column.|alt=]] |
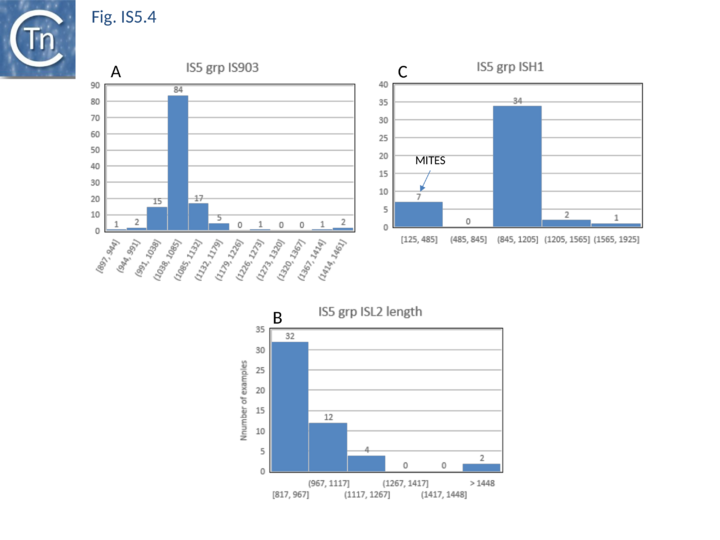
| − | [[Image:Fig. IS5.4.png|thumb|center| | + | [[Image:Fig. IS5.4.png|thumb|center|720x720px|'''Fig. IS5.4.''' Length (base pairs) distribution of IS''5'' family [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''] subgroups. The number of examples used in the sample is shown above each column.|alt=]] |
| − | There is a distant relationship, about 30% similarity, between IS''5'' and the Pif/Harbinger group of eukaryotic TE<ref><nowiki><pubmed>15020481</pubmed></nowiki></ref>. | + | There is a distant relationship, about 30% similarity, between [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] and the Pif/Harbinger group of eukaryotic TE<ref><nowiki><pubmed>15020481</pubmed></nowiki></ref>. |
=====Organization===== | =====Organization===== | ||
| − | Although the majority of members have a single Tpase orf, about 20% may express Tpase by [[wikipedia:Frameshift_mutation|frameshifting]] since it is distributed between two translation phases similar to most of the IS''427'' subgroup (82/116)<ref name=":1" />. In these cases if frameshifting indeed occurs the frameshifting signals appear more appropriate for a [[General Information/Transposase expression and activity#Programmed Transcriptional Frameshifting|programmed transcriptional realignment frameshift]] mechanism (PTR) rather than for classical [[General Information/Transposase expression and activity#Programmed Translational Frameshifting|translation frameshifting]] (PRF) since there are no obvious downstream enhancement signals<ref><nowiki><pubmed>21673094</pubmed></nowiki></ref>. | + | Although the majority of members have a single Tpase orf, about 20% may express Tpase by [[wikipedia:Frameshift_mutation|frameshifting]] since it is distributed between two translation phases similar to most of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''] subgroup (82/116)<ref name=":1" />. In these cases if frameshifting indeed occurs the frameshifting signals appear more appropriate for a [[General Information/Transposase expression and activity#Programmed Transcriptional Frameshifting|programmed transcriptional realignment frameshift]] mechanism (PTR) rather than for classical [[General Information/Transposase expression and activity#Programmed Translational Frameshifting|translation frameshifting]] (PRF) since there are no obvious downstream enhancement signals<ref><nowiki><pubmed>21673094</pubmed></nowiki></ref>. |
| − | Similar split reading frames have also been identified in several of the other subgroups: IS''1031'' (13/65 members); IS''L2'' (7/43); and few in the IS''5'' subgroup (7/149). There is no experimental evidence that these frameshift signals are functional but many of these IS are in multiple copies suggesting that the derivatives are active. In view of their diversity compared to families such as IS''3'', the subgroups will certainly be partitioned into additional groups as more ISs are identified. | + | Similar split reading frames have also been identified in several of the other subgroups: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''] (13/65 members); [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''] (7/43); and few in the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] subgroup (7/149). There is no experimental evidence that these frameshift signals are functional but many of these IS are in multiple copies suggesting that the derivatives are active. In view of their diversity compared to families such as [[IS Families/IS3 family|IS''3'']], the subgroups will certainly be partitioned into additional groups as more ISs are identified. |
| − | At present, the IS''903'' and the archaeal IS''H1'' subgroups whose IR are quite similar ([[:File:Fig. IS5.5.png|Fig.5.5]]) do not contain members with potential frameshifting. | + | At present, the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] and the archaeal [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''] subgroups whose '''IR''' are quite similar ([[:File:Fig. IS5.5.png|Fig.5.5]]) do not contain members with potential frameshifting. |
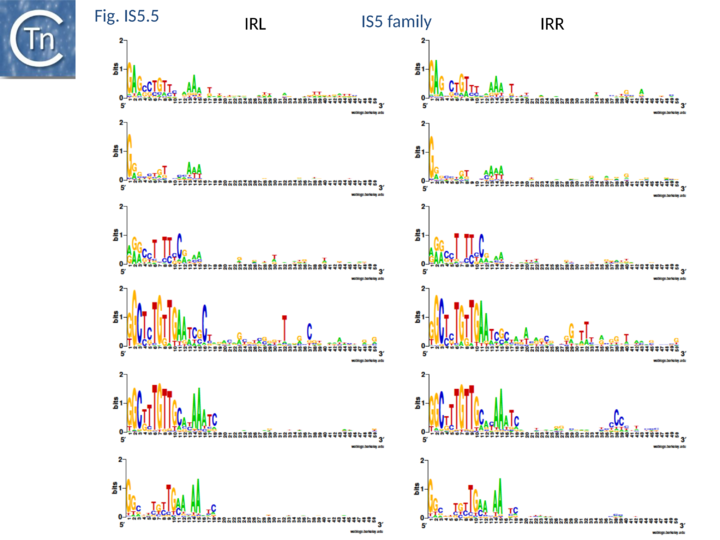
| − | [[Image:Fig. IS5.5.png|thumb|center| | + | [[Image:Fig. IS5.5.png|thumb|center|720x720px|'''Fig. IS5.5.''' '''[http://weblogo.threeplusone.com/ WebLogo] showing the most common ISH1 and IS''903'' ends.''' The left (IRL) and right IRR inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format. From top to bottom: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''], [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] subgroups.|alt=]] |
In addition to their Tpases and the presence or absence of potential frameshifting, a further distinction between these elements resides in their target specificities. | In addition to their Tpases and the presence or absence of potential frameshifting, a further distinction between these elements resides in their target specificities. | ||
| − | Certain IS''427'' subgroup members and IS''1182'' family members do not carry a termination codon for their Tpases but generate this on insertion into a specific target sequence, CTAG, which is duplicated on insertion. Other IS such as IS''1031'', duplicate a sequence TNA while others such IS''L2'' appear to duplicate ANT. | + | Certain [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''] subgroup members and IS''1182'' family members do not carry a termination codon for their Tpases but generate this on insertion into a specific target sequence, CTAG, which is duplicated on insertion. Other IS such as [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''], duplicate a sequence TNA while others such [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''] appear to duplicate ANT. |
| − | The lengths of the entire group range from 789 bp (IS''Mbu1'') to 1643 bp (IS''493''). The latter carries a second open reading frame upstream of the "Tpase" frame inessential for transposition<ref><nowiki><pubmed>1319378</pubmed></nowiki></ref>. IS''4811'' (Tn''4811''<ref><nowiki><pubmed>1332944</pubmed></nowiki></ref>, which is greater than 5kb, clearly contains a number of passenger genes including one with a consensus [[wikipedia:ATP-binding_motif|ATP/GTP-binding motif]]; an [[wikipedia:Oxidoreductase|oxidoreductase-like protein]]; and one related to bacterial transcription regulators of the [https://www.wikigenes.org/e/gene/e/1034955.html AraC family]. Another, IS''881'' from ''[[wikipedia:Streptomyces|Streptomyces]]'', is interrupted by a [[wikipedia:Group_II_intron|group II intron]]. | + | The lengths of the entire group range from 789 bp (e.g., [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISMbu1 IS''Mbu1'']) to 1643 bp (eg., [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS493 IS''493'']). The latter carries a second open reading frame upstream of the "Tpase" frame inessential for transposition<ref><nowiki><pubmed>1319378</pubmed></nowiki></ref>. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS4811 IS''4811''] (Tn''4811''<ref><nowiki><pubmed>1332944</pubmed></nowiki></ref>, which is greater than 5kb, clearly contains a number of passenger genes including one with a consensus [[wikipedia:ATP-binding_motif|ATP/GTP-binding motif]]; an [[wikipedia:Oxidoreductase|oxidoreductase-like protein]]; and one related to bacterial transcription regulators of the [https://www.wikigenes.org/e/gene/e/1034955.html AraC family]. Another, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS881 IS''881''] from ''[[wikipedia:Streptomyces|Streptomyces]]'', is interrupted by a [[wikipedia:Group_II_intron|group II intron]]. |
The major feature which defines this group is the similarities between their putative Tpases<ref><nowiki><pubmed>7934941</pubmed></nowiki></ref>. This includes the N2, N3 and C1 domains carried by the IS''4'' group<ref><nowiki><pubmed>7934941</pubmed></nowiki></ref>. However, IS''5'' family Tpases exhibit a spacing between the N3 and C1 domains of approximately 40 residues, a distance more consistent with the canonical DDE motif<ref name=":1" />. | The major feature which defines this group is the similarities between their putative Tpases<ref><nowiki><pubmed>7934941</pubmed></nowiki></ref>. This includes the N2, N3 and C1 domains carried by the IS''4'' group<ref><nowiki><pubmed>7934941</pubmed></nowiki></ref>. However, IS''5'' family Tpases exhibit a spacing between the N3 and C1 domains of approximately 40 residues, a distance more consistent with the canonical DDE motif<ref name=":1" />. | ||
| − | Analysis of the largely increased number of members generally confirms these subgroups. Members within each group also generate distinct DRs of similar lengths (IS''5'', 4 bp; IS''L2'', 2-3 bp; IS''1031'', 3-4 bp; IS''903'', 8-9 bp; and IS''427'', 2-3 bp). | + | Analysis of the largely increased number of members generally confirms these subgroups. Members within each group also generate distinct DRs of similar lengths ([https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''], 4 bp; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''], 2-3 bp; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''], 3-4 bp; [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''], 8-9 bp; and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''], 2-3 bp). |
| − | The IS''903'' and IS''H1'' subgroups have similar terminal IRs ([[:File:Fig. IS5.5.png|Fig.5.5]]) but appear distinct by correlation with the length of the target duplication and, to a lesser extent, by the typical length of the entire IS ([[:File:Fig. IS5.4.png|Fig.5.4]]). | + | The [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''] subgroups have similar terminal IRs ([[:File:Fig. IS5.5.png|Fig.5.5]]) but appear distinct by correlation with the length of the target duplication and, to a lesser extent, by the typical length of the entire IS ([[:File:Fig. IS5.4.png|Fig.5.4]]). |
| − | Several members exhibit GATC sites within their terminal 50 bp. This includes all members of the IS''903'' subgroup and many members of the IS''1031'' and IS''427'' subgroups. IS''903'' transposition activity has been shown to be modulated by Dam ''in vivo'' (cited in <ref><nowiki><pubmed>3000598</pubmed></nowiki></ref>). | + | Several members exhibit GATC sites within their terminal 50 bp. This includes all members of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] subgroup and many members of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''] subgroups. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposition activity has been shown to be modulated by Dam ''in vivo'' (cited in <ref><nowiki><pubmed>3000598</pubmed></nowiki></ref>). |
| − | A preferred target sequence, YTAR (often CTAG), is observed for two subgroups, IS''5'' and IS''427'', and for two members of the IS''L2'' group (IS''112'' and IS''1373'') in which either all four base pairs or the central TA are duplicated on insertion. | + | A preferred target sequence, YTAR (often CTAG), is observed for two subgroups, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS427 IS''427''], and for two members of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''] group (eg., [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS112 IS''112''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1373 IS''1373'']) in which either all four base pairs or the central TA are duplicated on insertion. |
| − | It is important to underline that, in many cases, the sequence of the original target site before insertion is not available. This can introduce ambiguities not only in estimating the number of duplicated target base pairs but also in defining the IRs. It is particularly important in several cases where the target repeat is symmetrical (e.g. CTAG) and where it is impossible to distinguish whether the element duplicates 2 or 4 bp and therefore to determine the exact ends of the element. Alignment of the ends of these elements in subgroups has permitted a number of ambiguities to be resolved. Members of the IS''L2'' group which generate 3 bp DRs exhibit a preference for ANT while those from the IS''1031'' group (which generate exclusively a 3 bp DR) exhibits a preference for insertion sites with the sequence TNA. Neither the small IS''H1'' group (8 bp DRs) nor the IS''903'' group (9 bp DRs) exhibit marked target specificity (see IS''903'' and also [[General Information/Target Choice|Target specificity]]). | + | It is important to underline that, in many cases, the sequence of the original target site before insertion is not available. This can introduce ambiguities not only in estimating the number of duplicated target base pairs but also in defining the '''IRs'''. It is particularly important in several cases where the target repeat is symmetrical (e.g. CTAG) and where it is impossible to distinguish whether the element duplicates 2 or 4 bp and therefore to determine the exact ends of the element. Alignment of the ends of these elements in subgroups has permitted a number of ambiguities to be resolved. Members of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISL2 IS''L2''] group which generate 3 bp DRs exhibit a preference for ANT while those from the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1031 IS''1031''] group (which generate exclusively a 3 bp '''DR''') exhibits a preference for insertion sites with the sequence TNA. Neither the small [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISH1 IS''H1''] group (8 bp DRs) nor the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] group (9 bp DRs) exhibit marked target specificity (see [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] and also [[General Information/Target Choice|Target specificity]]). |
| − | Only two of these elements, IS''5'' and IS''903'', have received significant attention. | + | Only two of these elements, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''], have received significant attention. |
=====IS''5'' group===== | =====IS''5'' group===== | ||
| − | In spite of the historical importance of IS5 in generating mutations, the published work concerning this element is largely directed to an understanding of its coding capacity and expression properties. IS5 carries one large orf, ins5A, spanning the entire element and shown to be essential for transposition IS5 (see <ref name=":0" />), and two small orfs (ins5B and 5C<ref><nowiki><pubmed>6269958</pubmed></nowiki></ref><ref><nowiki><pubmed>6269959</pubmed></nowiki></ref><ref><nowiki><pubmed>6281651</pubmed></nowiki></ref><ref><nowiki><pubmed>6327289</pubmed></nowiki></ref>, whose relevance to transposition remains to be demonstrated. Nothing is known about the transposition mechanism of this element. | + | In spite of the historical importance of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] in generating mutations, the published work concerning this element is largely directed to an understanding of its coding capacity and expression properties. [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] carries one large orf, ins5A, spanning the entire element and shown to be essential for transposition [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] (see <ref name=":0" />), and two small orfs (ins5B and 5C<ref><nowiki><pubmed>6269958</pubmed></nowiki></ref><ref><nowiki><pubmed>6269959</pubmed></nowiki></ref><ref><nowiki><pubmed>6281651</pubmed></nowiki></ref><ref><nowiki><pubmed>6327289</pubmed></nowiki></ref>, whose relevance to transposition remains to be demonstrated. Nothing is known about the transposition mechanism of this element. |
=====Mechanism IS''903''===== | =====Mechanism IS''903''===== | ||
| − | The only IS''5'' family member which transposition mechanism has been addressed at present is IS''903''. The ends of IS''903'' carry IRs of 18 bp which exhibit the typical two-domain organization<ref><nowiki><pubmed>2825175</pubmed></nowiki></ref> . Transposase has been shown to bind specifically to the ends using a region located in the amino-terminal portion of the protein<ref><nowiki><pubmed>1324175</pubmed></nowiki></ref><ref name=":2"><pubmed>9417930</pubmed> | + | The only [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS5 IS''5''] family member which transposition mechanism has been addressed at present is [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903'']. The ends of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] carry IRs of 18 bp which exhibit the typical two-domain organization<ref><nowiki><pubmed>2825175</pubmed></nowiki></ref> . Transposase has been shown to bind specifically to the ends using a region located in the amino-terminal portion of the protein<ref><nowiki><pubmed>1324175</pubmed></nowiki></ref><ref name=":2"><pubmed>9417930</pubmed> |
| + | |||
| + | </nowiki></ref>. In addition, a region possibly involved in the formation of higher order multimers has been identified and residues probably involved in catalysis have been pinpointed among the conserved residues in the catalytic DDE domain<ref name=":2" />. Insertion generates a 9 bp target duplication. | ||
| − | An elegant genetic analysis provided strong evidence that IS''903'' is not only capable of undergoing direct insertion but can also generate adjacent deletions in a duplicative manner. Moreover, point mutations in the terminal base pair of the IRs decrease overall transposition frequency but increase the frequency of [[wikipedia:Cointegrate|cointegrate formation]]<ref><nowiki><pubmed>10096085</pubmed></nowiki></ref>. Similarly, mutation of the first nucleotide flanking an IR also influences the level of [[wikipedia:Cointegrate|cointegrate formation]]<ref><nowiki><pubmed>11387225</pubmed></nowiki></ref>. The level of [[wikipedia:Cointegrate|cointegrate formation]] can also be increased by mutation of the Tpase. The molecular nature of these effects requires further investigation. | + | An elegant genetic analysis provided strong evidence that IS''903'' is not only capable of undergoing direct insertion but can also generate adjacent deletions in a duplicative manner. Moreover, point mutations in the terminal base pair of the '''IRs''' decrease overall transposition frequency but increase the frequency of [[wikipedia:Cointegrate|cointegrate formation]]<ref><nowiki><pubmed>10096085</pubmed></nowiki></ref>. Similarly, mutation of the first nucleotide flanking an IR also influences the level of [[wikipedia:Cointegrate|cointegrate formation]]<ref><nowiki><pubmed>11387225</pubmed></nowiki></ref>. The level of [[wikipedia:Cointegrate|cointegrate formation]] can also be increased by mutation of the Tpase. The molecular nature of these effects requires further investigation. |
| − | Factors affecting IS''903'' target site choice have been addressed in some detail. Initial studies<ref><nowiki><pubmed>9620951</pubmed></nowiki></ref> identified that insertion into the conjugative plasmid pOX38 showed no consensus in the 9 bp target duplication produced on insertion but the alignment of the target sequences indicated a preference for sites with symmetry on either side. A cloned copy of one native symmetric site into a second conjugative plasmid, pUB307, confirmed its attractiveness for insertion. More extensive studies provided a consensus symmetric target sequence which, when cloned into a target replicon, proved highly efficient<ref><nowiki><pubmed>11178901</pubmed></nowiki></ref>. The preferred target was a 21 bp palindrome cantered on the 9 bp target duplication. It could be dissected into: the 5 bp flanking sequences, the most important for site-specific insertion; the 7 bp palindromic core within the target duplication; the dinucleotide pair at the transposon-target junction; and the local DNA context. | + | Factors affecting [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] target site choice have been addressed in some detail. Initial studies<ref><nowiki><pubmed>9620951</pubmed></nowiki></ref> identified that insertion into the conjugative plasmid pOX38 showed no consensus in the 9 bp target duplication produced on insertion but the alignment of the target sequences indicated a preference for sites with symmetry on either side. A cloned copy of one native symmetric site into a second conjugative plasmid, pUB307, confirmed its attractiveness for insertion. More extensive studies provided a consensus symmetric target sequence which, when cloned into a target replicon, proved highly efficient<ref><nowiki><pubmed>11178901</pubmed></nowiki></ref>. The preferred target was a 21 bp palindrome cantered on the 9 bp target duplication. It could be dissected into: the 5 bp flanking sequences, the most important for site-specific insertion; the 7 bp palindromic core within the target duplication; the dinucleotide pair at the transposon-target junction; and the local DNA context. |
| − | Insertion into pUB307 itself showed a strong preference for a single orientation. By inverting either the vegetative (''oriV'') or transfer, ''oriT'', origins, it was concluded that orientation was determined by the direction of conjugative transfer. This of course implies that the ends of IS''903'' are not equivalent. It also implies, as is the case for Tn''7''<ref><nowiki><pubmed>8804309</pubmed></nowiki></ref><ref><nowiki><pubmed>11274058</pubmed></nowiki></ref><ref><nowiki><pubmed>11030337</pubmed></nowiki></ref><ref><nowiki><pubmed>26104363</pubmed></nowiki></ref><ref><nowiki><pubmed>19703395</pubmed></nowiki></ref> and members of the IS''200''/IS''608'' family<ref><nowiki><pubmed>26350330</pubmed></nowiki></ref><ref><nowiki><pubmed>27466393</pubmed></nowiki></ref><ref><nowiki><pubmed>20691900</pubmed></nowiki></ref>, that transposition targets [[wikipedia:DNA_replication#Replication_fork|replication forks]]. | + | Insertion into pUB307 itself showed a strong preference for a single orientation. By inverting either the vegetative (''oriV'') or transfer, ''oriT'', origins, it was concluded that orientation was determined by the direction of conjugative transfer. This of course implies that the ends of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] are not equivalent. It also implies, as is the case for [http://tncentral.ncc.unesp.br/cgi-bin/tn_report.pl?id=Tn7-NC_002525 Tn''7'']<ref><nowiki><pubmed>8804309</pubmed></nowiki></ref><ref><nowiki><pubmed>11274058</pubmed></nowiki></ref><ref><nowiki><pubmed>11030337</pubmed></nowiki></ref><ref><nowiki><pubmed>26104363</pubmed></nowiki></ref><ref><nowiki><pubmed>19703395</pubmed></nowiki></ref> and members of the [[IS Families/IS200-IS605 family|IS''200''/IS''608'' family]]<ref><nowiki><pubmed>26350330</pubmed></nowiki></ref><ref><nowiki><pubmed>27466393</pubmed></nowiki></ref><ref><nowiki><pubmed>20691900</pubmed></nowiki></ref>, that transposition targets [[wikipedia:DNA_replication#Replication_fork|replication forks]]. |
| − | The requirement the most abundant nucleoid proteins in transposition<ref><nowiki><pubmed>15130124</pubmed></nowiki></ref>. Most notably, H-NS was required for efficient transposition. Similar results were obtained for IS''10'' and Tn''522'' suggesting a more general role for H-NS in bacterial transposition. H-NS exerts its effect on target capture: IS''903''. Targeting preferences in the E. coli chromosome were dramatically altered in the absence of H-NS. | + | The requirement the most abundant nucleoid proteins in transposition<ref><nowiki><pubmed>15130124</pubmed></nowiki></ref>. Most notably, [[wikipedia:Histone-like_nucleoid-structuring_protein|H-NS]] was required for efficient transposition. Similar results were obtained for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10''] and Tn''522'' suggesting a more general role for [[wikipedia:Histone-like_nucleoid-structuring_protein|H-NS]] in bacterial transposition. [[wikipedia:Histone-like_nucleoid-structuring_protein|H-NS]] exerts its effect on target capture: [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903'']. Targeting preferences in the E. coli chromosome were dramatically altered in the absence of [[wikipedia:Histone-like_nucleoid-structuring_protein|H-NS]]. |
| − | Several other host mutants were identified exhibiting a unique population pattern<ref name=":3"><pubmed>15968071</pubmed> | + | Several other host mutants were identified exhibiting a unique population pattern<ref name=":3"><pubmed>15968071</pubmed></nowiki></ref>: a ring phenotype with predominant papillae located just inside the edge of the colony, implying a spatial triggering of transposition within the. These mutants were found to be in ''pur'' genes, whose products are involved in purine biosynthesis. The genetic evidence was consistent with a requirement for GTP in [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposition. These observations suggest that transposition occurs in later stages of colony growth. Transposition may occur within the colony edge in response to either a gradient of exogenous purines across the colony and may also reflect the developmental stage of the cells. |
| − | IS''903'' transposase like those of a variety of other IS, exhibits a strong preference for action in cis: complementation of defective transposons in trans occurs at less than 1% <ref name=":3" />. Transposition is extremely sensitive to the distance between the 3' end of the transposase gene and the nearest transposon IR. Insertion of 1 kb of DNA reduces transposition to 1-2%. There is a strong correlation between the stability of transposase and its ability to act in trans. wild-type transposase has a half-life of about 3 min. Fusion with [[wikipedia:Alpha-galactosidase|α-galactosidase]] stabilizes the protein and results in an increase in its capacity to act in trans. A similar effect was noted in a lon mutant strain where trans activity was increased by a factor of 10-100. Further studies identified a class of transposase mutants specifically enhanced in trans activity and reduced in cis activity without increasing the overall transposition frequency. This was correlated with an increase in transposase half-life compared to the wildtype Derbyshire<ref><nowiki><pubmed>8898394</pubmed></nowiki></ref>. A second class of mutants with enhanced cis activity resulted in increased levels of transposase expression (as for IS''10''<ref><nowiki><pubmed>8412678</pubmed></nowiki></ref>). | + | [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS903 IS''903''] transposase like those of a variety of other IS, exhibits a strong preference for action in cis: complementation of defective transposons in trans occurs at less than 1% <ref name=":3" />. Transposition is extremely sensitive to the distance between the 3' end of the transposase gene and the nearest transposon '''IR'''. Insertion of 1 kb of DNA reduces transposition to 1-2%. There is a strong correlation between the stability of transposase and its ability to act in trans. wild-type transposase has a half-life of about 3 min. Fusion with [[wikipedia:Alpha-galactosidase|α-galactosidase]] stabilizes the protein and results in an increase in its capacity to act in trans. A similar effect was noted in a lon mutant strain where trans activity was increased by a factor of 10-100. Further studies identified a class of transposase mutants specifically enhanced in trans activity and reduced in '''cis''' activity without increasing the overall transposition frequency. This was correlated with an increase in transposase half-life compared to the wildtype Derbyshire<ref><nowiki><pubmed>8898394</pubmed></nowiki></ref>. A second class of mutants with enhanced cis activity resulted in increased levels of transposase expression (as for [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS10R IS''10'']<ref><nowiki><pubmed>8412678</pubmed></nowiki></ref>). |
====IS''1182''==== | ====IS''1182''==== | ||
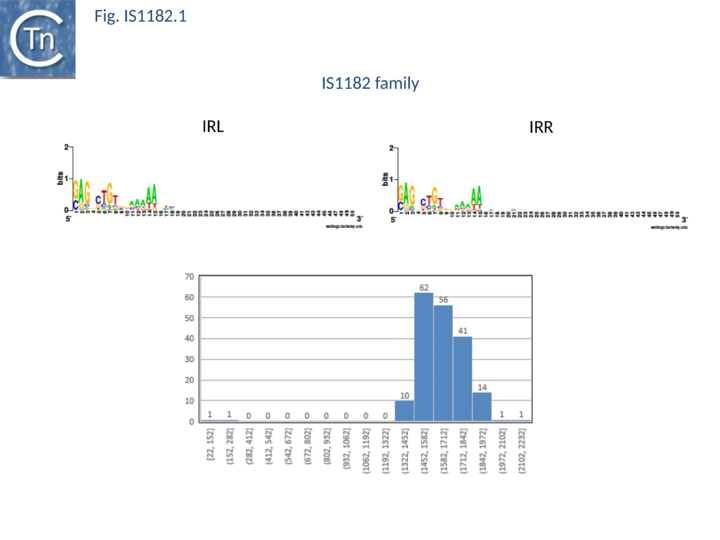
| − | IS''1182'' family members exhibit a diverse set of target specificities. Some duplicate 4 bp. These are of two types: those specific for CTAG and those that show no apparent target sequence specificity. Yet others target palindromic sequences. These are also of different types: some insert at the 3’ foot of a stem-loop and duplicate the entire structure while others insert 3’ of the loop and simply duplicate the loop ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier], [https://www.france-bioinformatique.fr/en/users/gourbeyre-edith E. Gourbeyre] and [https://scholar.google.fr/citations?user=r8TYgVEAAAAJ&hl=fr M. Chandler], unpublished) ([[:File:Fig. IS1182.1.png|Fig.IS1182.1]]). | + | [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1182 IS''1182''] family members exhibit a diverse set of target specificities. Some duplicate 4 bp. These are of two types: those specific for CTAG and those that show no apparent target sequence specificity. Yet others target palindromic sequences. These are also of different types: some insert at the 3’ foot of a stem-loop and duplicate the entire structure while others insert 3’ of the loop and simply duplicate the loop ([https://scholar.google.fr/citations?user=WHAtfqcAAAAJ&hl=fr P. Siguier], [https://www.france-bioinformatique.fr/en/users/gourbeyre-edith E. Gourbeyre] and [https://scholar.google.fr/citations?user=r8TYgVEAAAAJ&hl=fr M. Chandler], unpublished) ([[:File:Fig. IS1182.1.png|Fig.IS1182.1]]). |
| − | [[Image:Fig. IS1182.1.png|thumb|center| | + | [[Image:Fig. IS1182.1.png|thumb|center|720x720px|'''Fig. IS1182.1.''' '''The IS''1182'' family's main characteristics. Top:''' The left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format. '''Bottom''': distribution of IS length (base pairs) [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1182 IS''1182''] family members. The number of examples used in the sample is shown above each column. |alt=]] |
====IS''Dol1'' group (ISNCY)==== | ====IS''Dol1'' group (ISNCY)==== | ||
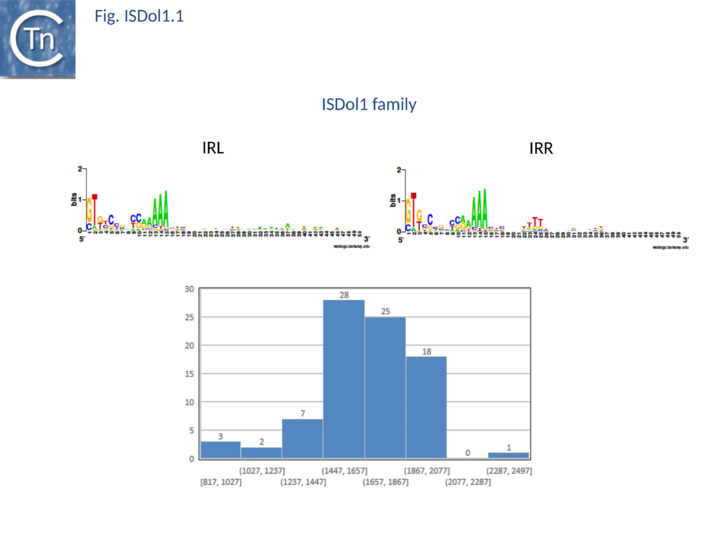
| − | Another small group, IS''Dol1'', with 58 members from a large number of bacterial species has emerged from the ISNCY “orphan” group. Members have a length of between 1600-1900 bp ([[:File:Fig. ISDol1.1.png|Fig.ISDol.1]]) and generate DRs of 6-7bp. | + | Another small group, [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISDol1 IS''Dol1''], with 58 members from a large number of bacterial species has emerged from the ISNCY “orphan” group. Members have a length of between 1600-1900 bp ([[:File:Fig. ISDol1.1.png|Fig.ISDol.1]]) and generate DRs of 6-7bp. |
| − | [[Image:Fig. ISDol1.1.png|thumb|center| | + | [[Image:Fig. ISDol1.1.png|thumb|center|720x720px|'''Fig. ISDol1.1.''' '''The IS''1182'' family's main characteristics.''' '''Top''': The left ('''IRL''') and right '''IRR''' inverted terminal repeats are shown in [http://weblogo.threeplusone.com/ WebLogo] format. '''Bottom''': distribution of IS length (base pairs) [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1182 IS''1182''] family members. The number of examples used in the sample is shown above each column. |alt=]] |
==Bibliography== | ==Bibliography== | ||
<references /> | <references /> | ||
Revision as of 12:47, 11 August 2021
Contents
IS5 family
Original Identification
IS5 was originally isolated as an insertion into the immunity region of bacteriophage lambda and subsequently found as a cause of mutation in a number of E. coli genes[1][2][3][4]. Together with IS1, it was also identified as an activator (by insertion) of expression of the usually cryptic beta-glucosidase gene of E. coli[5][6][7][8][9][10].
Presence in Compound Transposons
Several members are associated with compound transposons. These include IS903 and IS602, which form part of the kanamycin resistance transposons Tn903[11], and Tn602[12] respectively, and ISVa1 and ISVa2 which form part of a transposon carrying iron transport genes[13].
Distribution
The IS5 family, like the IS4 family, is also a relatively heterogeneous group which now requires reanalysis. It also includes sequences from both eubacteria and the archaea. There are now a large number of identified members of the IS5 family (>550 members) and of a closely related IS1182 family (>150 members) which have allowed a more detailed analysis and a separation into various subgroups and families. The IS5 family is partitioned into 6 subgroups: IS5, IS903, ISL2, ISH1, IS1031 and IS427[14] (Table Characteristics of IS families; Fig.5.1). Some of these may prove to be emerging families. Members of the IS5 subgroup appear to be composed of two groups with different lengths: one of 1060-1300 bp and a second of 1460-1610 (Fig.5.2 A).
Diversity
The transposases of these are also of different lengths (Fig.5.2 B) and transposase length is correlated with that of the IS. The lengths of the IS1013 subgroup are between ~900 and ~1200 bp with the majority between 103 and 1090 bp (Fig.5.3), those of the IS427 group are between 800 and 1070 bp in length with most having lengths in the range of 810 900 bp (Fig.5.3). Members of the IS903 subgroup are generally about 1030-1090 bp long (Fig.5.4 A ), those of the ISH1 subgroup are about 850 - 1200 bp long (note that this subgroup includes a number of Miniature Inverted repeat Transposable Elements (MITES) (Fig.5.4 B) and ISL2 members are 820 to 1260 bp long with a majority of about 820-970 bp (Fig.5.4 C). There are a large number of additional IS5 family members whose attribution to subgroups has yet to be established.
There is a distant relationship, about 30% similarity, between IS5 and the Pif/Harbinger group of eukaryotic TE[15].
Organization
Although the majority of members have a single Tpase orf, about 20% may express Tpase by frameshifting since it is distributed between two translation phases similar to most of the IS427 subgroup (82/116)[14]. In these cases if frameshifting indeed occurs the frameshifting signals appear more appropriate for a programmed transcriptional realignment frameshift mechanism (PTR) rather than for classical translation frameshifting (PRF) since there are no obvious downstream enhancement signals[16].
Similar split reading frames have also been identified in several of the other subgroups: IS1031 (13/65 members); ISL2 (7/43); and few in the IS5 subgroup (7/149). There is no experimental evidence that these frameshift signals are functional but many of these IS are in multiple copies suggesting that the derivatives are active. In view of their diversity compared to families such as IS3, the subgroups will certainly be partitioned into additional groups as more ISs are identified.
At present, the IS903 and the archaeal ISH1 subgroups whose IR are quite similar (Fig.5.5) do not contain members with potential frameshifting.
In addition to their Tpases and the presence or absence of potential frameshifting, a further distinction between these elements resides in their target specificities.
Certain IS427 subgroup members and IS1182 family members do not carry a termination codon for their Tpases but generate this on insertion into a specific target sequence, CTAG, which is duplicated on insertion. Other IS such as IS1031, duplicate a sequence TNA while others such ISL2 appear to duplicate ANT.
The lengths of the entire group range from 789 bp (e.g., ISMbu1) to 1643 bp (eg., IS493). The latter carries a second open reading frame upstream of the "Tpase" frame inessential for transposition[17]. IS4811 (Tn4811[18], which is greater than 5kb, clearly contains a number of passenger genes including one with a consensus ATP/GTP-binding motif; an oxidoreductase-like protein; and one related to bacterial transcription regulators of the AraC family. Another, IS881 from Streptomyces, is interrupted by a group II intron.
The major feature which defines this group is the similarities between their putative Tpases[19]. This includes the N2, N3 and C1 domains carried by the IS4 group[20]. However, IS5 family Tpases exhibit a spacing between the N3 and C1 domains of approximately 40 residues, a distance more consistent with the canonical DDE motif[14].
Analysis of the largely increased number of members generally confirms these subgroups. Members within each group also generate distinct DRs of similar lengths (IS5, 4 bp; ISL2, 2-3 bp; IS1031, 3-4 bp; IS903, 8-9 bp; and IS427, 2-3 bp).
The IS903 and ISH1 subgroups have similar terminal IRs (Fig.5.5) but appear distinct by correlation with the length of the target duplication and, to a lesser extent, by the typical length of the entire IS (Fig.5.4).
Several members exhibit GATC sites within their terminal 50 bp. This includes all members of the IS903 subgroup and many members of the IS1031 and IS427 subgroups. IS903 transposition activity has been shown to be modulated by Dam in vivo (cited in [21]).
A preferred target sequence, YTAR (often CTAG), is observed for two subgroups, IS5 and IS427, and for two members of the ISL2 group (eg., IS112 and IS1373) in which either all four base pairs or the central TA are duplicated on insertion.
It is important to underline that, in many cases, the sequence of the original target site before insertion is not available. This can introduce ambiguities not only in estimating the number of duplicated target base pairs but also in defining the IRs. It is particularly important in several cases where the target repeat is symmetrical (e.g. CTAG) and where it is impossible to distinguish whether the element duplicates 2 or 4 bp and therefore to determine the exact ends of the element. Alignment of the ends of these elements in subgroups has permitted a number of ambiguities to be resolved. Members of the ISL2 group which generate 3 bp DRs exhibit a preference for ANT while those from the IS1031 group (which generate exclusively a 3 bp DR) exhibits a preference for insertion sites with the sequence TNA. Neither the small ISH1 group (8 bp DRs) nor the IS903 group (9 bp DRs) exhibit marked target specificity (see IS903 and also Target specificity). Only two of these elements, IS5 and IS903, have received significant attention.
IS5 group
In spite of the historical importance of IS5 in generating mutations, the published work concerning this element is largely directed to an understanding of its coding capacity and expression properties. IS5 carries one large orf, ins5A, spanning the entire element and shown to be essential for transposition IS5 (see [7]), and two small orfs (ins5B and 5C[22][23][24][25], whose relevance to transposition remains to be demonstrated. Nothing is known about the transposition mechanism of this element.
Mechanism IS903
The only IS5 family member which transposition mechanism has been addressed at present is IS903. The ends of IS903 carry IRs of 18 bp which exhibit the typical two-domain organization[26] . Transposase has been shown to bind specifically to the ends using a region located in the amino-terminal portion of the protein[27][28]. In addition, a region possibly involved in the formation of higher order multimers has been identified and residues probably involved in catalysis have been pinpointed among the conserved residues in the catalytic DDE domain[28]. Insertion generates a 9 bp target duplication.
An elegant genetic analysis provided strong evidence that IS903 is not only capable of undergoing direct insertion but can also generate adjacent deletions in a duplicative manner. Moreover, point mutations in the terminal base pair of the IRs decrease overall transposition frequency but increase the frequency of cointegrate formation[29]. Similarly, mutation of the first nucleotide flanking an IR also influences the level of cointegrate formation[30]. The level of cointegrate formation can also be increased by mutation of the Tpase. The molecular nature of these effects requires further investigation.
Factors affecting IS903 target site choice have been addressed in some detail. Initial studies[31] identified that insertion into the conjugative plasmid pOX38 showed no consensus in the 9 bp target duplication produced on insertion but the alignment of the target sequences indicated a preference for sites with symmetry on either side. A cloned copy of one native symmetric site into a second conjugative plasmid, pUB307, confirmed its attractiveness for insertion. More extensive studies provided a consensus symmetric target sequence which, when cloned into a target replicon, proved highly efficient[32]. The preferred target was a 21 bp palindrome cantered on the 9 bp target duplication. It could be dissected into: the 5 bp flanking sequences, the most important for site-specific insertion; the 7 bp palindromic core within the target duplication; the dinucleotide pair at the transposon-target junction; and the local DNA context.
Insertion into pUB307 itself showed a strong preference for a single orientation. By inverting either the vegetative (oriV) or transfer, oriT, origins, it was concluded that orientation was determined by the direction of conjugative transfer. This of course implies that the ends of IS903 are not equivalent. It also implies, as is the case for Tn7[33][34][35][36][37] and members of the IS200/IS608 family[38][39][40], that transposition targets replication forks.
The requirement the most abundant nucleoid proteins in transposition[41]. Most notably, H-NS was required for efficient transposition. Similar results were obtained for IS10 and Tn522 suggesting a more general role for H-NS in bacterial transposition. H-NS exerts its effect on target capture: IS903. Targeting preferences in the E. coli chromosome were dramatically altered in the absence of H-NS.
Several other host mutants were identified exhibiting a unique population pattern[42]: a ring phenotype with predominant papillae located just inside the edge of the colony, implying a spatial triggering of transposition within the. These mutants were found to be in pur genes, whose products are involved in purine biosynthesis. The genetic evidence was consistent with a requirement for GTP in IS903 transposition. These observations suggest that transposition occurs in later stages of colony growth. Transposition may occur within the colony edge in response to either a gradient of exogenous purines across the colony and may also reflect the developmental stage of the cells.
IS903 transposase like those of a variety of other IS, exhibits a strong preference for action in cis: complementation of defective transposons in trans occurs at less than 1% [42]. Transposition is extremely sensitive to the distance between the 3' end of the transposase gene and the nearest transposon IR. Insertion of 1 kb of DNA reduces transposition to 1-2%. There is a strong correlation between the stability of transposase and its ability to act in trans. wild-type transposase has a half-life of about 3 min. Fusion with α-galactosidase stabilizes the protein and results in an increase in its capacity to act in trans. A similar effect was noted in a lon mutant strain where trans activity was increased by a factor of 10-100. Further studies identified a class of transposase mutants specifically enhanced in trans activity and reduced in cis activity without increasing the overall transposition frequency. This was correlated with an increase in transposase half-life compared to the wildtype Derbyshire[43]. A second class of mutants with enhanced cis activity resulted in increased levels of transposase expression (as for IS10[44]).
IS1182
IS1182 family members exhibit a diverse set of target specificities. Some duplicate 4 bp. These are of two types: those specific for CTAG and those that show no apparent target sequence specificity. Yet others target palindromic sequences. These are also of different types: some insert at the 3’ foot of a stem-loop and duplicate the entire structure while others insert 3’ of the loop and simply duplicate the loop (P. Siguier, E. Gourbeyre and M. Chandler, unpublished) (Fig.IS1182.1).
ISDol1 group (ISNCY)
Another small group, ISDol1, with 58 members from a large number of bacterial species has emerged from the ISNCY “orphan” group. Members have a length of between 1600-1900 bp (Fig.ISDol.1) and generate DRs of 6-7bp.
Bibliography
- ↑ <pubmed>4432374</pubmed>
- ↑ <pubmed>353507</pubmed>
- ↑ <pubmed>84614</pubmed>
- ↑ <pubmed>641012</pubmed>
- ↑ <pubmed>6270569</pubmed>
- ↑ <pubmed>3034860</pubmed>
- ↑ 7.0 7.1 </nowiki>
- ↑ <pubmed>2846278</pubmed>
- ↑ <pubmed>7781607</pubmed>
- ↑ <pubmed>8710516</pubmed>
- ↑ <pubmed>6261245</pubmed>
- ↑ <pubmed>2819910</pubmed>
- ↑ <pubmed>7568465</pubmed>
- ↑ 14.0 14.1 14.2 </nowiki>
- ↑ <pubmed>15020481</pubmed>
- ↑ <pubmed>21673094</pubmed>
- ↑ <pubmed>1319378</pubmed>
- ↑ <pubmed>1332944</pubmed>
- ↑ <pubmed>7934941</pubmed>
- ↑ <pubmed>7934941</pubmed>
- ↑ <pubmed>3000598</pubmed>
- ↑ <pubmed>6269958</pubmed>
- ↑ <pubmed>6269959</pubmed>
- ↑ <pubmed>6281651</pubmed>
- ↑ <pubmed>6327289</pubmed>
- ↑ <pubmed>2825175</pubmed>
- ↑ <pubmed>1324175</pubmed>
- ↑ 28.0 28.1 Tavakoli NP, DeVost J, Derbyshire KM . Defining functional regions of the IS903 transposase. - J Mol Biol: 1997 Dec 12, 274(4);491-504 [PubMed:9417930] [DOI] </nowiki>
- ↑ <pubmed>10096085</pubmed>
- ↑ <pubmed>11387225</pubmed>
- ↑ <pubmed>9620951</pubmed>
- ↑ <pubmed>11178901</pubmed>
- ↑ <pubmed>8804309</pubmed>
- ↑ <pubmed>11274058</pubmed>
- ↑ <pubmed>11030337</pubmed>
- ↑ <pubmed>26104363</pubmed>
- ↑ <pubmed>19703395</pubmed>
- ↑ <pubmed>26350330</pubmed>
- ↑ <pubmed>27466393</pubmed>
- ↑ <pubmed>20691900</pubmed>
- ↑ <pubmed>15130124</pubmed>
- ↑ 42.0 42.1 </nowiki>
- ↑ <pubmed>8898394</pubmed>
- ↑ <pubmed>8412678</pubmed>