IS Families/IS91-ISCR families
Contents
History
IS91 was identified in the early 1980s in a number of hemolytic plasmids of different incompatibility groups [1] and characterized in the alpha haemolytic Escherichia coli plasmid, pSU233, as a 1.85 kb insertion sequence which could transpose sequentially to a variety of other plasmids including pACYC184, R388, and pBR322. These original studies identified a majority of simple insertions but also a few percents of apparent cointegrates [2]. A number of related IS have been identified and two, IS801 from a multi-antibiotic resistance Escherichia coli plasmid pUB2380 [3] and IS1294 from a Pseudomonas syringae plasmid [4], like IS91, have also been shown to be active in transposition. More recently, it has been suggested that another group of related sequences, the ISCR (for IS with Common Regions), are instrumental in transmitting antibiotic resistance genes [5][6]. IS91 family members have related TEs in eukaryotes, the helitrons [7][8][9].
Organization
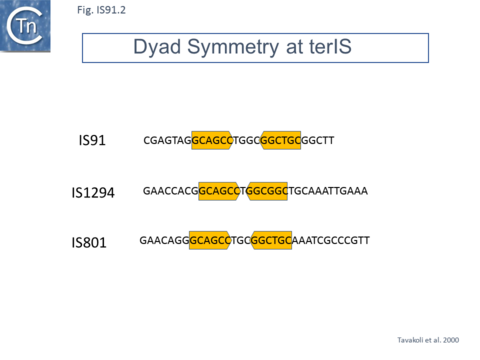
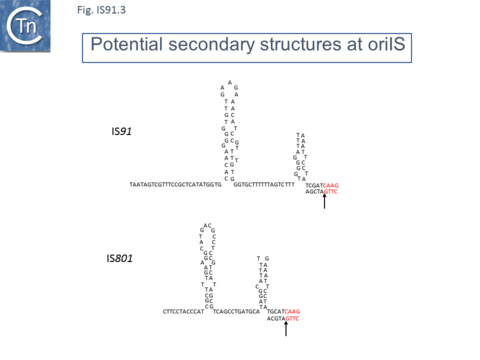
IS91 itself (Fig.IS91.1) includes two imperfect terminals 8 base pair inverted repeats (5'-TCGAGTAGG….CCTATCGA-3'). The ends also include a variety of sequences with dyad symmetry which might form secondary structures or simply are the recognition signals for transposase binding. The left end, terIS (see Mechanism below) carries short hexanucleotide inverted repeats also observed in both IS801 and IS1294 while neither element exhibits imperfect terminal IR [4][10](Fig.IS91.2) . The right end of IS91 and of IS801, oriIS, shows a more complex pattern of dyad symmetry (Fig.IS91.3) , although IS1294 only possesses a single short sequence with dyad symmetry at this end.
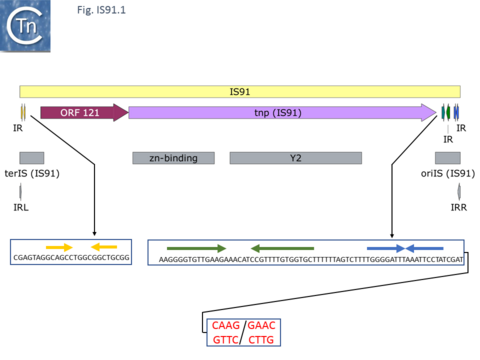
Unlike the majority of insertion sequences and transpososns, IS91 family members do not generate direct target repeats ( target site duplications) on insertion [11]. IS91, IS801 and IS1294 appear to insert specifically 5' to 5'-GAAC or 5'-CAAG tetranucleotide with the same relative orientation to the target sequence [4][12][13] (Fig.IS91.1 and Fig.IS91.3). The DNA sequence of IS91 showed a long orf, the transposase with a cysteine-rich N-terminal region probably forming a zinc finger (Fig.IS91.4a) and a C-terminal catalytic domain comprising a HUH triad together with two tyrosine residues (Y2) (Fig.IS91.4b). Other members of the family also exhibit these characteristic motifs (Fig.IS91.4c). The second orf of unknown function is located upstream and its termination codon overlaps the initiation codon of the transposase indicating that expression of the two proteins may be translationally coupled. Insertional mutagenesis of the longer orf using Tn1732 demonstrated that it was essential for transposition [14] whereas insertion into the upstream orf had no effect on transposition frequency but appears to impact the choice of the target [15]. This orf is absent from both IS801 and IS1294 as well as in all other IS91 family members in ISfinder.
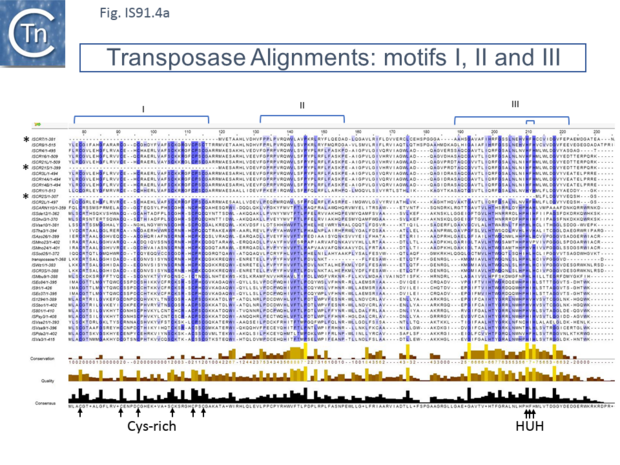
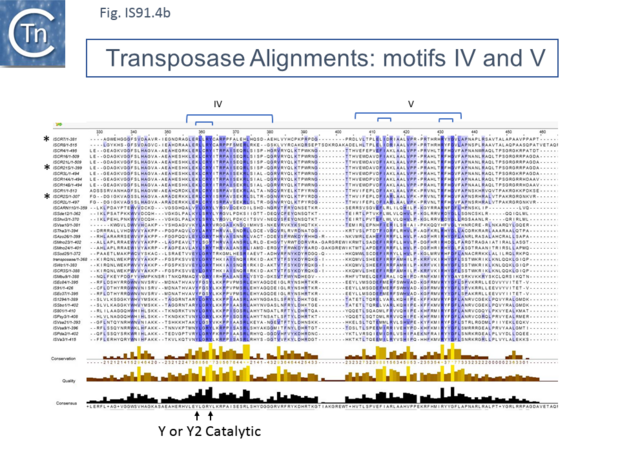
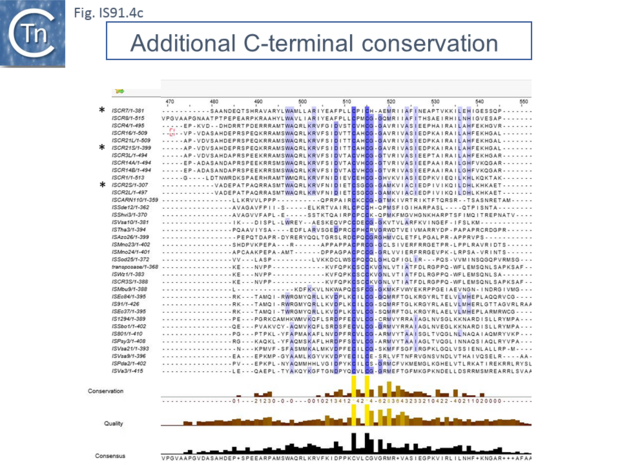
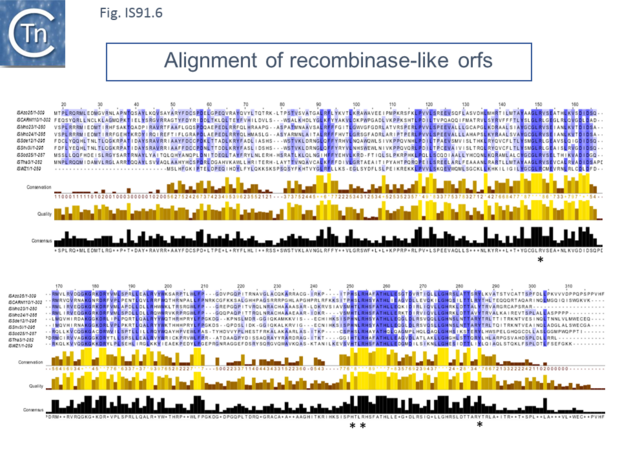
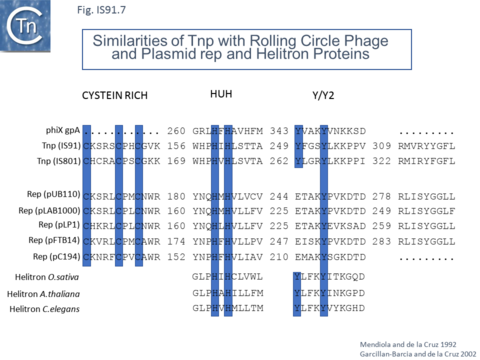
However, in the limited number of IS91 family members catalogued in ISfinder, several appear to include an additional orf, unrelated to that in IS91, either upstream (ISAzo26, ISCARN110, ISMno23, ISSde12, ISShvi3, ISSod25 and ISWz1) or down-stream (ISMno24 and ISTha3) of the transposase gene. BLAST analysis shows the product of this orf to be related to tyrosine recombinases (Fig.IS91.6). Alignments of the associated transposases (Fig.IS91.4a, b and c, and Fig.IS91.5) fall into three groups: related IS91 family members (including IS91, IS801 and IS1294), those which have been classified as ISCR (see below) and those that also carry the putative tyrosine recombinase. Those IS which carry the second integrase orf and the ISCR transposases carry only a single catalytic Y residue in the HUH transposase (Fig.IS91.4b). The transposases encoded by these IS are related to those of eukaryotic helitrons [16] and to the rep proteins which initiate rolling circle replication in some plasmids and ssDNA viruses such as that of AAV (Fig.IS91.7). They are also related to, yet topologically distinct from, relaxases of conjugal plasmids due a circular permutation of their domains. A comparison of various members of the HUH family of endonucleases is shown in Fig.7.5 (see [17]). Several include 5' to 3' helicase domains which might facilitate DNA unwinding during the replication/transfer process. The IS91 and ISCR transposases do not, suggesting that they might rely on host enzymes for this function.
Mutagenesis of the IS91 transposase demonstrated that both Y residues (Y249 and Y253) are essential for transposition in vivo [18].
Distribution
A survey of the sequence databases using IS91 transposase as a the query revealed very similar transposases present in a range of bacterial genera including Bergeyella, Fusobacterium, Rhizobium, Shewanella, Pseudomonas, Klebsiella, Shigella and Escherichia [10] and a number of related orfs in Mesorhizobium, Pseudomonas, Vibrio and Salmonella [19]. These include both chromosomal and plasmid-carried copies. BLAST analysis of the present public protein sequence databases (June 2020) showed the transposases of the family to be very widely spread. While the identification of the transposase is straightforward, the definition of the entire IS at the DNA level is more problematic due to the general absence of terminal inverted repeats and of target site duplications therefore only afew full length IS have been identified.
Mechanism
The similarity to rolling circle replication proteins both plasmid and viral suggested that IS91 and other family members transpose using a novel rolling circle transposition mechanism [4][16]. This idea was reinforced by in vivo studies on its transposition behavior. In addition to its specific insertion at 5'-GAAC (GTTC) or 5'-CAAG (CTTG) target sequences, IS91 insertion was found to be orientation-specific: the right end is inserted adjacent 5' to the target sequences as shown in Fig.IS91.1 (note the DNA strand polarity in this figure is such that cleavage would occur on the bottom strand). It was also shown that the flanking tetranucleotide target was necessary for further transposition along with an 81 base pair sequence within the right end, oriIS, while the left end was dispensable.
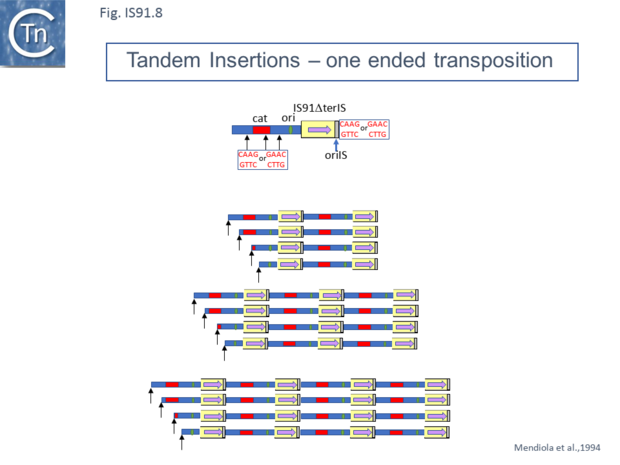
Mutants in which terIS had been deleted were found to retain transposition activity and generated insertions of tandem multimers of the donor plasmid (Fig.IS91.8 and Fig.IS91.9) with one transposon/target junction precisely at the oriIS end and the other variable carrying a different length of plasmid sequence but flanked by a tetranucleotide target sequence [20]. Analysis of transposition products using a plasmid transposon donor carrying an IS91 copy with a deleted left end (terIS) in mating out assay identified fusions of the donor and target plasmids which occurred at roughly the same frequency as cointegrates with a wild-type IS. Examples of two, three and 4 tandem copies of the inserted donor plasmid were observed (Fig.IS91.8). Since each of the variable ends of the insert (left in Fig.IS91.8) terminated with a characteristic GTTC or CTTG tetranucleotide resident in the donor plasmid, this is consistent with a rolling circle type transposition mechanism where the process initiates by nicking of the the oriIS tetranucleotide copy and, in the absence of the terIS sequence, terminates inefficiently by recognition of the secondary target tetranucleotide within the donor plasmid DNA.
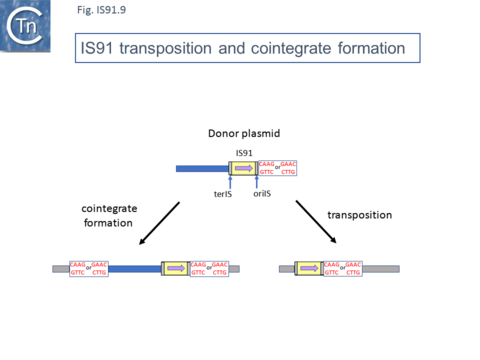
Normal transposition results in the insertion of the IS so that oriIS abuts the conserved target tetranucleotide with terIS defining the other boundary. The low level (2%) of cointegrates observed in early studies [2][11] suggests that they are generated by the type of one ended transposition observed in the terIS deleted IS mutants (Fig.IS91.9). The relationship between these two types of event at the molecular level is unclear at the moment. Clearly the signal, terIS, recognized in normal transposition is different from that recognized in cointegrate formation or “one-ended” transposition (the tetranucleotide sequence).
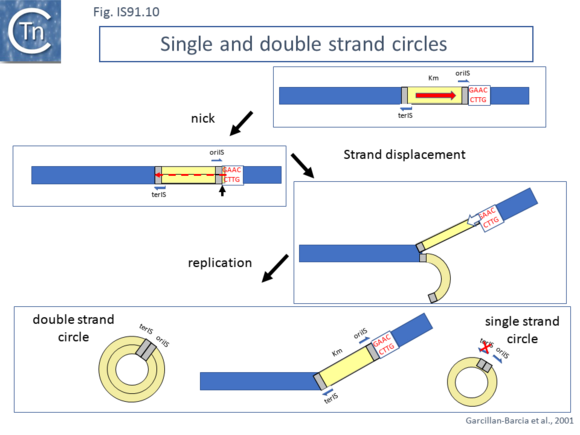
Studies using an artificial IS91 derivative in which oriIS and terIS flank a selectable kanamycin resistance gene carried by a plasmid with an independently inducible IS91 transposase gene revealed that induction of transposase expression resulted in the production of two novel DNA species. One was sensitive to single-strand nuclease S1 and which showed complementarity to an oligonucleotide expected to hybridize with oriIS on the strand thought to be displaced and transferred but not to a terIS-specific oligonucleotide with complementarity to the opposite strand (Fig.IS91.10) [18]. The other hybridized to both oriIS and terIS oligonucleotides. PCR amplification using appropriate oligonucleotides and each of the new DNA species and DNA sequencing showed that both carried a junction in which oriIS and terIS were abutted. Circle formation depends on an intact transposase Y2 motif. This suggested a circle formation mechanism which was dependent on an presence of both active site tyrosines of the transposase. However, it is not yet clear whether circles either in the ssDNA or dsDNA forms are intermediates in the IS91 transposition pathway [18].
Transposition models
The present model(s) for rolling circle transposition is shown in Fig.12.2 and Fig.12.3. The first involves an initiation event at one IS end, polarized transfer of the IS strand into a target molecule, and termination at the second end Fig.12.2 [10][19]. An alternative model involving transposon circle formation is shown in Fig.12.3. This model is attractive since it would liberate the transposon circle intermediate to locating a target site following circle formation rather than requiring target engagement during the replicative transposition process as does the first model (Fig.12.2).
Transposition is postulated to initiate at IRR as a result of transposase-mediated cleavage (one of the active site tyrosines generates a 5’-phosphotyrosine born with the transposase), and the 3′-OH generated in the donor molecule would act as a primer for DNA replication while one transposon DNA strand is peeled off. Transfer of the donor sequence to a target is driven by replication in the donor, during which displacement of the active transposon strand would be driven by leading-strand replication (in the absence of a transposase associated helicase domain). Transfer of an ssDNA IS91 copy into the target DNA is accomplished when ter‑IS is reached and cleaved. This occurs after the replication of the entire transposon. A second transposase-catalyzed event is thought to result in strand-transfer by nicking the 3’-end of the transposon and joining it to the 5’-end of a target site.
Termination fails at a high frequency (1-2% of all insertions), resulting in so-called one-ended transposition, as it results in the insertion of additional flanking donor-plasmid genes 3′ to the IRL sequence. The dsDNA circles could result from replication restart or from the extension of a trapped Okazaki fragment on the excised ssDNA circle. The relationship between transposition of IS91 and replication of the donor and target replicons is not known. It is also not clear what role, if any, the circular transposition intermediates play.
Note that IS91 insertion is reminiscent of that of members of the IS200/IS605 family[21] which have been shown to transpose using a single stranded circular intermediate by a mechanism called “peel and paste”. These also insert with one end next to a specific tetranucleotide and this, like that of IS91, the target is also required for further transposition.
Flanking gene acquisition is thought to occur when the termination mechanism fails and rolling circle transposition extends into neighboring DNA where it may encounter a second surrogate end [10]. These types of mobile element may prove to play an important role in the assembly and transmission of multiple antibiotic resistance [5][22].
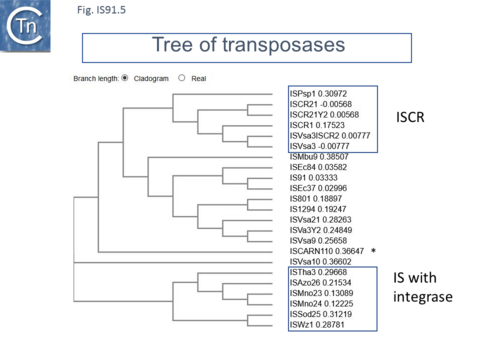
ISCR
More recently, a group of related elements, ISCR (IS with a "Common Region") was described (reviewed in [5][23][24]). The CR is an orf which resembles the IS91 family transposases [17] previously called orf513 [25]. A major feature of ISCR elements is that they are associated with a variety of antibiotic resistance genes. It is possible that ICRs can facilitate the movement of neighboring resistance genes by a mechanism similar to one-ended transposition exhibited by IS91 (Fig.IS91.9). An alternative view may be that the major impact of ISCR elements is via homologous recombination exchange rather than transposition per se [5][26]. The fact that ISCR elements ISCR3, ISCR4, ISCR14, and ISCR16, which show between 75% and 97% identity in their transposase orf, and are located downstream from a groE-like orf has led to the suggestion that they may all be descended from a common ancestor [27]. Since their sequence environment is identical, this further suggests that they are relatively inactive in transposition. There is as yet no formal demonstration that these transpose.
ISCR1
ISCR1 is almost exclusively associated with integrons and the end which, according to the IS91/IS801/IS1294 model should carry terIS, and is always located downstream from a qacH-sul1 gene pair (specifying fluoroquinolone and sulfonamide resistance respectively) with an upstream integron recombination site (Fig.IS91.11). As shown by Toleman et al. [5][24] all members they identified from GenBank using the ISCR1 DNA sequence as a query exhibited this feature although there was variation further upstream due to loss and capture of integron cassettes. This upstream sequence identity makes it problematic to define the terIS sequence. However, if ISCR1 was responsible for ABR gene movement using the “one-ended” transposition mechanism of IS91 family elements, it is the sequences flanking the terIS end that should show diversity. Toleman et al.[24] suggested that one-ended transposition indeed occurs but that the secondary signal which provokes termination of transposition occurs upstream of the intI gene (Fig.IS91.11). Variation in the flanking DNA sequence does occur but this is flanking the oriIS end. Conservation of the ISCR1 sequence ends with GTGGTTTATACTTCCTATACCC in all examples [24] (Fig.IS91.11). It is not clear from these data whether there is a tetranucleotide target sequence because this would require the DNA sequence of an empty site that is not available. Given that, for IS91, this tetranucleotide is important for transposition, that data may actually indicate that the copies of ISCR1 identified are inactive.
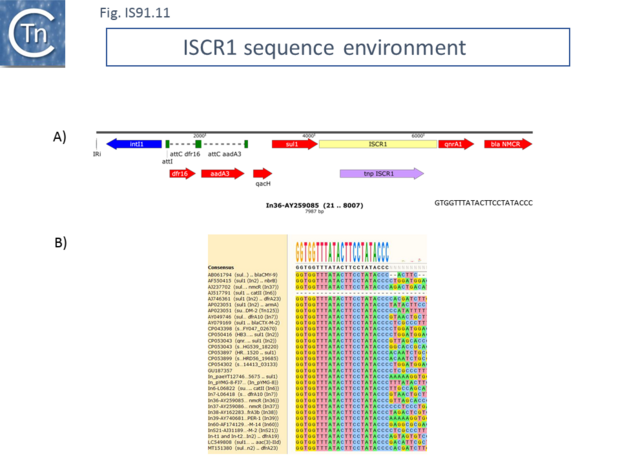
ISCR2
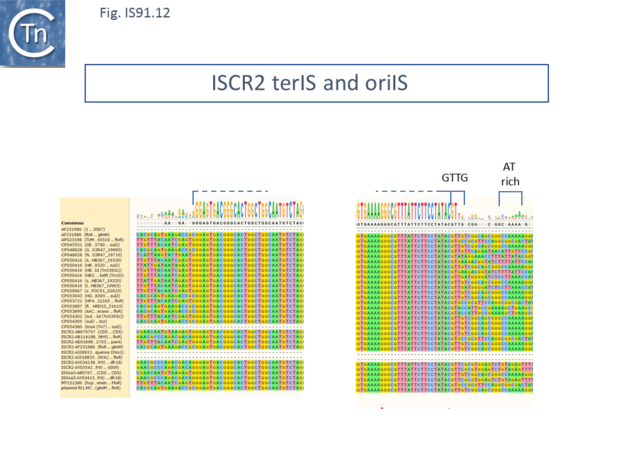
ISCR2 is cataloged in ISfinder as ISVsa3. However, further analysis indicated that all copies included a transposase with an N-terminal extension located outside and upstream of the IS DNA sequence. This carries the important Cysteine-rich zinc-binding domain and the HUH motifs of the canonical IS91 family transposases. ISCR2 has proven easier to define because of sequence diversification upstream (flanking terIS) and downstream (flanking oriIS) (Fig.IS91.12) and now encompasses the full-length transposase orf. ISCR2 exhibits a reasonably robust tetranucleotide at its oriIS end of GTTG (17/28) and there is an A/T-rich region located slightly downstream. ISCR2 there possesses all the characteristics of a complete IS91 family insertion sequence.
Little is known concerning the other ISCR (3 to 12) [5][24]. ISCR8 has also been cataloged in ISfinder as ISPsp1 and ISCR21, like ISVsa3 (ISCR2), include a transposase with an N-terminal extension located outside and upstream of the IS DNA sequence which carries the Cysteine-rich domain.
Helitrons
Interestingly related transposase proteins were also observed in a variety of eukaryotic organisms including plants and in the worm Caenorhabditis elegans [7] and some mammals such as bats. They can be identified, many as extinct fragments in the genomes of members of all eukaryotic kingdom [8][9][28][29][30]. Like IS91 family members, Helitron transposition is often associated with the capture and mobilization of host genomic fragments, resulting in the dissemination of genomic regulatory elements [28][31]. Unlike those of IS91 family members, their transposases possess a 5' to 3' helicase domain. downstream of the HUH endonuclease domain.
Progress had been made in understanding the mechanism of helitron movement using reconstituted transposon from the little brown bat, M. lucifugus, throwing light on how IS91 family members might transpose. Since there were no known active helitron copies, a functional copy, called Helraiser was reconstructed based on consensus sequences [32]. Its transposase (1,496 amino acids) exhibits large and unique N-terminal domain and a catalytic core with an HUH motif with two active site Tyr residues as do IS91 family transposases. This is preceded by an N-terminal eukaryote-specific nuclear localization-like signal and followed by a long (600-aa) helicase domain characteristic of the SF1b superfamily of DNA helicases, and carrying sequence motifs of the eukaryotic Pif1 helicases [32]. The transposase orf is bounded by left and right terminal sequences (LTS and RTS) terminating in conserved 5’-TC/CTAG-3’ which are characteristic of the Helibat1 helitron family [28]. Like IS91, RTS exhibits a 19bp palindrome just upstream.
Helraiser was able to form covalently closed transposon circles, just as does IS91 [18]. The sequence of the Helraiser circle junction revealed that the left end (LTS) 5′-TC dinucleotide was covalently joined to the CTAG-3′ tetranucleotide of the right end (RTS). In addition, plasmid molecules containing the cloned junction obtained by propagation in E.coli, underwent transposase-dependent integration when co-transfected into HeLa cells. The DNA sequence of 10 independent insertion junctions indicated that all had inserted into an AT dinucleotide, a general characteristic of Helitrons. Moreover, not only could the Helraiser transposase mobilize structures with its own ends, but it was also capable of mobilizing the cloned ends of two of three naturally occurring defective Helibat family members.
Mutation of the HUH motif or either of the two tyrosine residues (Y727 and Y731) in the catalytic domain or a Walker A box or arginine finger in the helicase domain reduced transposition to undetectable levels. It was also shown that the HUH motif and Y731 were required in vitro for cleavage of single-strand oligonucleotides representing the Helraiser ends. This study also investigated the sequences at each transposon end required for transposition in a similar way to the approach used for IS91 [20]. Deletion analysis showed than an intact LTS was required suggesting that it might be equivalent to oriIS of IS91 family members, while RTS was not strictly required although removal of the sequence with dyad symmetry or the entire RTS reduced activity. This, again, is similar to IS91 and would facilitate the transduction of neighboring genes.
Further studies addressing the nature of donor, target, and transposon intermediates [33] in human cells showed a requirement for double-stranded transposon donor molecules, although only one of the two transposon strands is used. Moreover, donor sites could be used multiple times implying that the transposon undergoes replication at the donor site after a single strand is “peeled off”. Double strand transposon circles were also identified and these behave as transposon donors in the presence of transposase. Strand-specific cleavage and transfer were also demonstrated in an in vitro system in this study.
Presumably, many of these mechanistic observations will also apply to the IS91 family of prokaryotic insertion sequences.
Bibliography
- ↑ <pubmed>PMC220263</pubmed>
- ↑ 2.0 2.1 </nowiki>
- ↑ <pubmed>1646375</pubmed>
- ↑ 4.0 4.1 4.2 4.3 </nowiki>
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 </nowiki>
- ↑ <pubmed>19763428</pubmed>
- ↑ 7.0 7.1 </nowiki>
- ↑ 8.0 8.1 Thomas J, Pritham EJ . Helitrons, the Eukaryotic Rolling-circle Transposable Elements. - Microbiol Spectr: 2015 Aug, 3(4); [PubMed:26350323] [DOI] </nowiki>
- ↑ 9.0 9.1 </nowiki>
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 Garcillán-Barcia MP, Bernales I, Mendiola MV, De La Cruz F. IS91 Rolling-Circle Transposition. In: Craig NL, Lambowitz AM, Craigie R, Gellert M, editors. Mobile DNA II. American Society of Microbiology; 2002. p. 891–904
- ↑ 11.0 11.1 Diaz-Aroca E, Mendiola MV, Zabala JC, de la Cruz F . Transposition of IS91 does not generate a target duplication. - J Bacteriol: 1987 Jan, 169(1);442-3 [PubMed:3025186] [DOI] </nowiki>
- ↑ <pubmed>2552258</pubmed>
- ↑ <pubmed>9870703</pubmed>
- ↑ <pubmed>PMC206431</pubmed>
- ↑ <pubmed>10411740</pubmed>
- ↑ 16.0 16.1 </nowiki>
- ↑ 17.0 17.1 </nowiki>
- ↑ 18.0 18.1 18.2 18.3 </nowiki>
- ↑ 19.0 19.1 Garcillán-Barcia MP, de la Cruz F . Distribution of IS91 family insertion sequences in bacterial genomes: evolutionary implications. - FEMS Microbiol Ecol: 2002 Nov 1, 42(2);303-13 [PubMed:19709290] [DOI] </nowiki>
- ↑ 20.0 20.1 </nowiki>
- ↑ <pubmed>26350330</pubmed>
- ↑ <pubmed>21729108</pubmed>
- ↑ <pubmed>PMC2916298</pubmed>
- ↑ 24.0 24.1 24.2 24.3 24.4 Toleman MA, Bennett PM, Walsh TR . Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. - J Antimicrob Chemother: 2006 Jul, 58(1);1-6 [PubMed:16751201] [DOI] </nowiki>
- ↑ <pubmed>PMC149023</pubmed>
- ↑ <pubmed>PMC127246</pubmed>
- ↑ <pubmed>PMC2565877</pubmed>
- ↑ 28.0 28.1 28.2 </nowiki>
- ↑ <pubmed>PMC2785235</pubmed>
- ↑ <pubmed>PMC2997563</pubmed>
- ↑ <pubmed>PMC4224331</pubmed>
- ↑ 32.0 32.1 </nowiki>
- ↑ <pubmed>PMC5876387</pubmed>