IS Families/IS30 family
Contents
General features and Original Identification
IS30 was identified as an insertion into the phage P1 genome[1][2].
Distribution
The IS30 family currently comprises more than 100 members from 94 members from nearly 80 bacterial species and an example, ISC1041, has also been found in the Archaea[3]. IS30-like Tpases have also been found as integral parts of certain Integrative Conjugative Elements (ICE) from Staphylococcus aureus (MRSA)[4]. An open reading frame found originally in a Spiroplasma citri phage (ΦSc1) but since identified also in the Spiroplasma citri genome is distantly related to the family.
Organization
IS30 family members have lengths of between 1027 bp (e.g. IS1070) and 1636 bp (e.g. ISCg2) with a single orf of between 293 and 398 codons spanning almost the entire length. The termination codon is generally very close to the right end. Alignment of the putative Tpases reveals a well-conserved DD(33)E motif.
Family members can activate neighboring genes by creation of a hybrid promoter on insertion next to a -10 promoter element[5][6][7][8].
The Tpases show several well-conserved regions. One, in the N-terminus includes a potential helix–turn–helix (HTH) motif, which, in the case of IS30, is responsible for IR binding[9][10]. Another in the C-terminal region contains the DD(33)E motif (Fig. IS30.1).
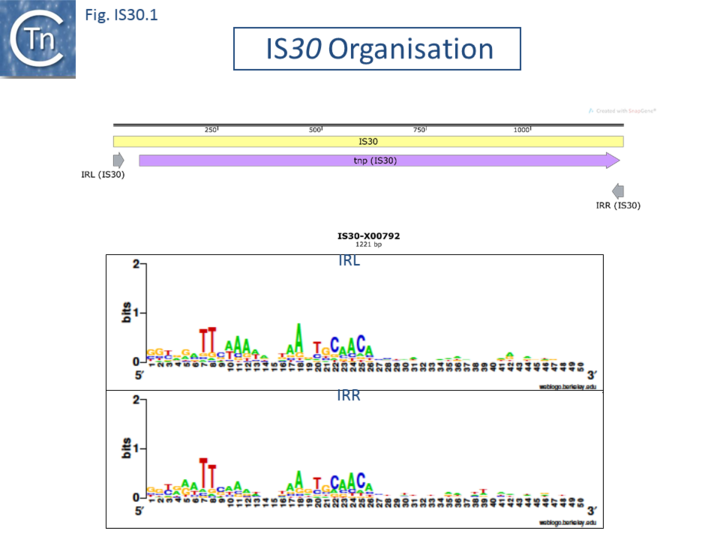
The terminal IRs are in the range of 20-30 bp and exhibit significantly conserved sequence signatures (Fig. IS30.1). The tips of the elements, as defined by the various authors, show significant sequence variation, although in most elements the IR has not been experimentally confirmed. Where it has been determined, 2 bp or 3 bp direct target repeats are generated on insertion, but there are several exceptions in which the DR is between 12 and 32 bp. [e.g. IS1630, IS1470, IS658, ISApl1, ISL7, ISLpl1[10]]. One member of this family, IS1630 (1377bp) from Mycoplasma fermentans is flanked by long but variable DRs of between 19 and 26bp[11].
Presence in Compound Transposons
IS30 is known to function in a compound transposon structure[12] and various IS30 family members also have been identified as part of composite transposons. In particular, Tn6330 which is composed of two copies of ISApl1 flanking the colistin resistance gene, mcr-1[13][14][15].
IS30
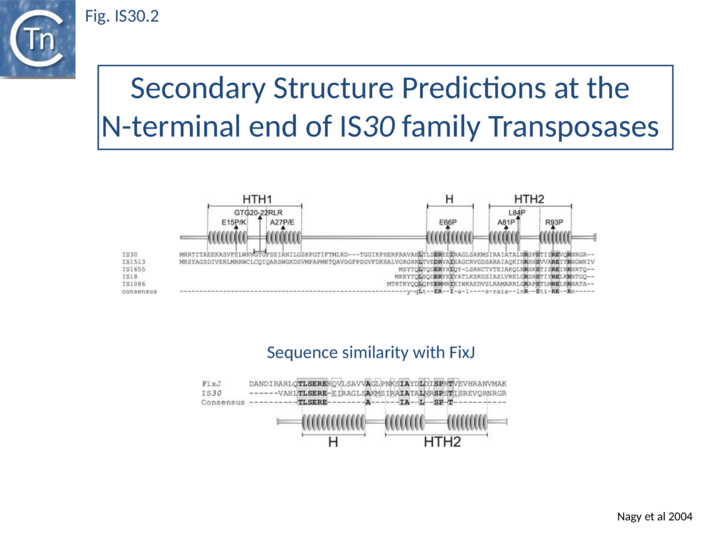
Of the different members of this family, IS30 is the best characterized. It is 1221 bp long with 26 bp terminal IRs and mediates several types of DNA rearrangements including simple insertion, cointegrate formation, deletion and inversion[2][16][17]. It shows pronounced insertion specificity and generates DRs of 2 bp. It terminates with the dinucleotide 5'-CA-3'[18] although many other members of the family do not. The single long orf is preceded by a relatively weak promoter (p30A) capable of driving Tpase expression with its -35 hexamer within IRL[19] and contains a weak internal transcription terminator. The transposase is predicted to include two N-terminal HTH motifs (Fig. IS30.2) and show significant homology to FixJ, a Sinorhizobium meliloti transcriptional regulator[10]. Premature transcription termination at this site could lead to the formation of Tpase molecules truncated at their C-terminal end carrying the catalytic DDE motif. Since the specific DNA binding domain which recognizes the terminal IRs is located in the N-terminal third of the protein[9], the potential truncated derivatives may lack the catalytic domain and regulate transposition activity by competition or interaction with a full-length Tpase. This arrangement may play the same role at the transcriptional level as does frameshifting at the translational level for certain other elements.
A second weak promoter (p30C) located on the opposite strand upstream of a short orf has been detected[19] and it has recently been shown that a small 150 nucleotide transcript driven from this promoter and contained entirely within the Tpase transcript acts as an antisense control by reducing translation of the Tpase mRNA[20].
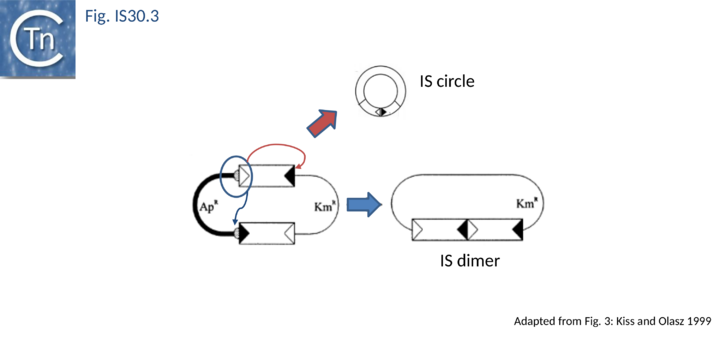
The founding member of the family, IS30, is also the best characterized at the mechanistic level[9][16][21][22][23][24][25][26] and an in vitro transposition system has been developed[27]. This 1221 bp long Escherichia coli element belongs to a growing class of ISs known to transpose through an intermediate formed by abutting the IRs, Donor Primed Transposon Replication. Here the IR are separated by 2 bp[5][16][23]. Such an IR junction can be created by formation of a dimer of two directly repeated IS copies or by the formation of transposon circles. Both IS mini-circles and dimers have been observed. IR–IR junctions have also been detected in some other IS30 family members such as IS18[7], IS4351[28] and IS1470[29]. A structure in which two IS30 ends are linked by a single strand bridge (forming a figure of eight structure on a circular plasmid), has been identified[27].
As in the case of IS21 and IS3 families, the assembly of abutted IRL and IRR copies create a strong promoter. In a tandem dimer of the element, this may result in increased expression of the Tpase from the downstream copy[5]. The IRL-IRR junction is also recombinationally unstable. It promotes transposition reactions at high frequency in vivo [16] presumably in a similar way to the equivalent junction of IS21 (see "IS21 family"). The activity of the junction appears maximal when the spacer between the abutted ends is 2 bp (the length of target duplications introduced when IS30 inserts). Lower activities are obtained with 1 and 3 bp spacers while little or no activity is observed for 0 bp or >3bp (T. Naas and W. Arber, pers. comm.). Strong evidence has recently been presented indicating that donor plasmid dimerization is followed by interaction of one appropriately oriented end from each IS copy to form an abutted IS intermediate (see Fig. IS30.3). The IS junction in this intermediate is highly active in transposition and plays an important role in transposition of IS30[23].
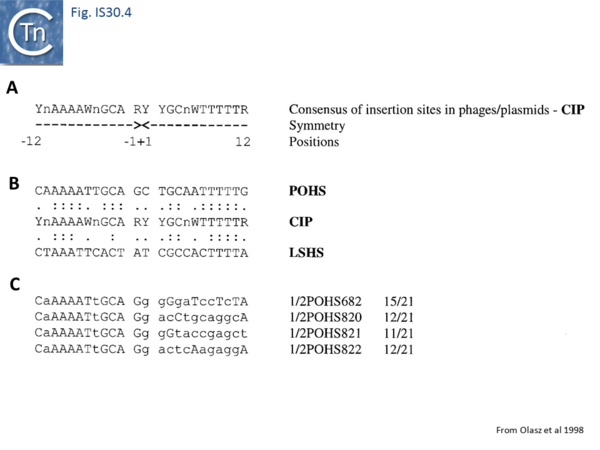
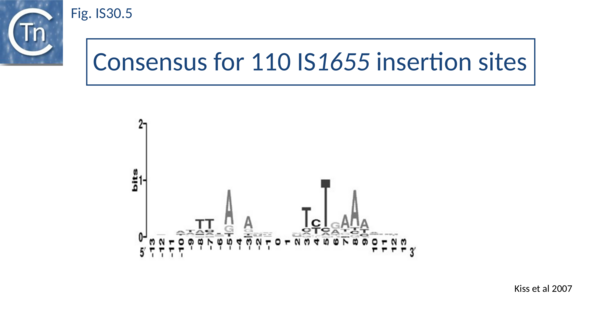
Like IS21[30] and IS911[31], IS30 shows a preference for insertion next to nucleotide sequences resembling its ends[22]. Furthermore, this region has been delimited approximately to the Tpase binding site. In addition, preferential insertion into a site in bacteriophage l (LHS) and P1 (OHS) has been observed both in E.coli [16] and Salmonella[32]. These sequences are somewhat degenerate palindromes characterized by a 24 bp symmetric consensus[33] (Fig. IS30.4). The sequence relationship between these various hotspots has yet to be explored. A similar type of insertion specificity was observed for IS1655 from Neisseria meningitis[26] (Fig. IS30.5). Another IS30 family member, ISCg2 from Corynebacterium glutamicum, has also been shown to insert into a relatively well defined sequence which exhibits some palindromic symmetry[34] while yet another, IS1630 from Mycoplasma fermentans, appears to show a preference for rather long target with extensive palindromic sequences[11].
ISApl1
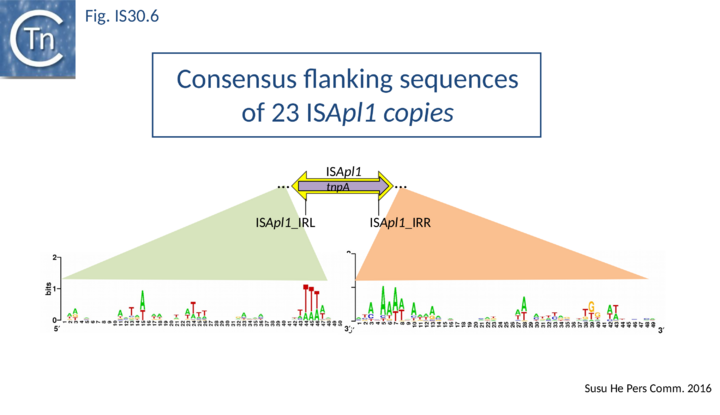
ISApl1 plays an important role in transmission of resistance to the last resort antibiotic, colistin [35][36][37][38][39][40][41]. It was originally identified in Actinobacillus pleuropneumoniae from pigs[42] and has typical IS30 family characteristics: 1070 bp long with a 924 bp transposase orf and flanked by 27 bp inverted repeats. Four copies occurred in the original AP76 strain, one of which interrupts the apxIVA toxin gene. It was also reported to be present in certain field isolates but not in the type strain. All four insertions generate a two base pair DR and occur in an AT rich region with some palindrome-like characteristics and a central GC rich region (see Fig. IS30.6) similar to that observed for IS1665[26]. Its transposase is very similar to that of IS30, especially around the active site.
PCR analysis demonstrated the presence of a junction fragment in which IRL and IRR are separated by a GG dinucleotide. This is the DR observed at the ISApl1 insertion site in A. pleuropneumoniae downstream of a bla gene but is not present at the other three insertion sites in this strain. The presence of this junction fragment suggests that ISApl1, like IS30 and IS1665, occurs using a circular DNA intermediate during transposition.
Following the initial report in 2016 of resistance to a last resort antibiotic, the polymyxin colistin, by a transmissible plasmid-associated mcr-1 gene [36], it was rapidly recognized (within the year) that the gene, which encodes a Phosphoethanolamine Transferase[36], was present in Enterobacteriaceae from five continents: Asia, Europe, Africa, South America, and North America (reviewed in [43]). Based on the analysis of 77 mcr-1-containing sequences (of which 19 were too fragmented to be useful) representing every unique sequence in NCBI as of August 2016, a common 2,607-bp DNA segment was identified that in many cases is flanked on one or both ends by ISApl1[14]. This fragment included the mcr-1 gene together with a truncated copy of a gene called pap2. This analysis led to the proposal that mcr-1 was transmitted by an ISApl1 composite transposon, later called Tn6330[15] which has, in some cases, subsequently lost one or both ISApl1 copies. Analysis of Tn6330 transposition has identified the expected transposition intermediate[44]: a transposon circle which carries a pair of abutted IS copies (as shown for IS30[23]; Fig. IS30.3). A second structure found in the study, a circular form with only a single IS copy may be equivalent to one of the transposition related deletion products described for IS30 [23] or simply by homologous recombination between the pair of directly repeated ISApl1. This was also observed in another study[15].
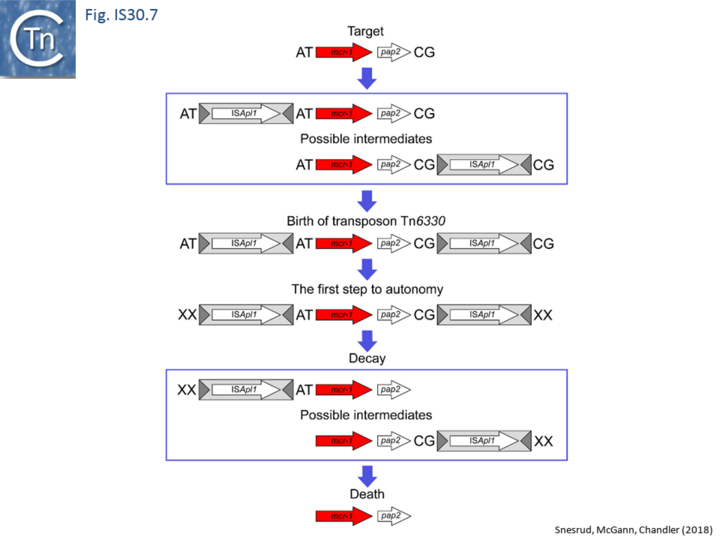
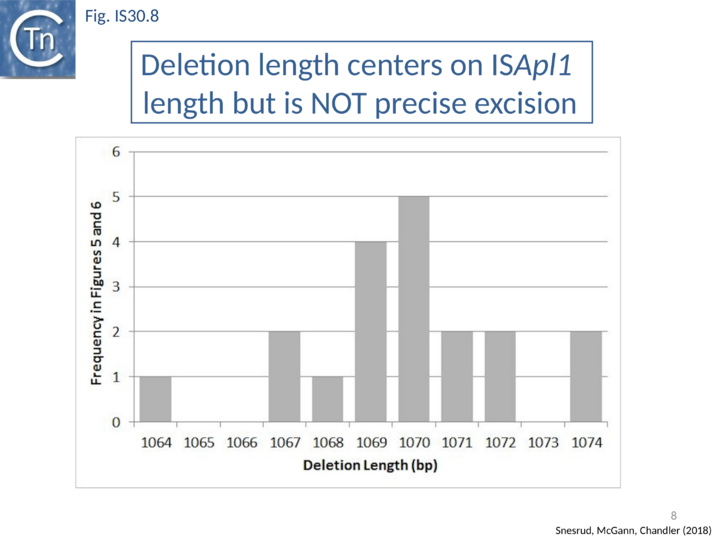
Further analysis with an enlarged database [13] provided further support for this and resulted in a model for the mobilization of mcr-1 from the chromosome of a species closely resembling Moraxella sp. MSG13-C03 by insertion of ISApl1 copies upstream and downstream (Fig. IS30.7), transmission by transposition to a target plasmid followed by interbacterial transfer and acquisition by additional conjugative plasmids. The sites of insertion were as expected for ISApl1 insertion sites (Fig. IS30.6), an observation which has been extensively confirmed[44]. The full transposon was then proposed to decay by deletion of the downstream and/or upstream ISApl1 copies leaving Tn6330 derivatives carrying a single downstream or upstream IS copy or devoid of both IS copies (Fig. IS30.7). Based on the transposition mechanism of IS30 family members, and on the observation that, although the deletion endpoints were variable from case to case, the length of the deletion was remarkably constant (Fig. IS30.8) representing the exact IS length, it was further proposed that IS deletion occurred by frequent abortive IS transposition (Fig. IS30.9 and Fig. IS30.10). IS30 family transposition appears to occur by a copy-out-paste-in mechanism which involves replication (Fig. IS30.9 left). In the deletion model, strand-switching between micro homologies during the replication step (Fig. IS30.9 right) would lead to the “looping out” of the IS rather than replication through to the opposite end. This could lead to removal of the IS copy during further plasmid replication [13]. This model is supported by data obtained several decades ago in which similar loss of IS30 was shown to occur with relatively high frequency and involve microhomologies[45]. Unfortunately, a more recent study that analyzed the stability of Tn6330 did not directly address the loss of flanking ISApl1 or the necessity of transposition enzymes or intact terminal IRs [15].
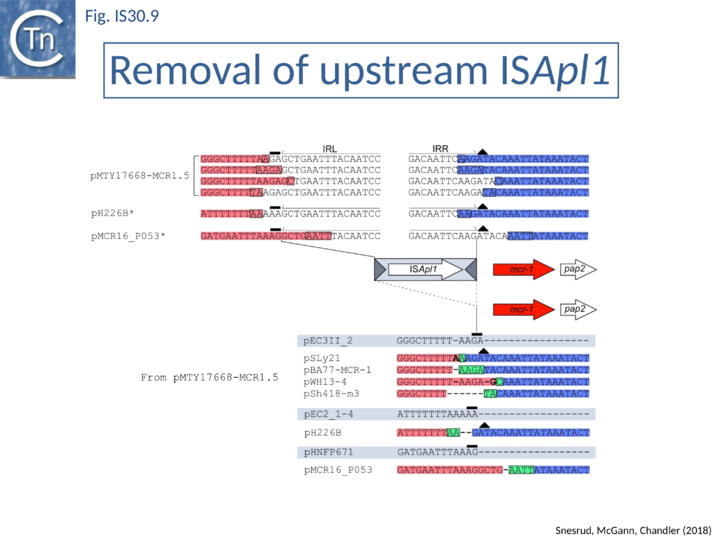
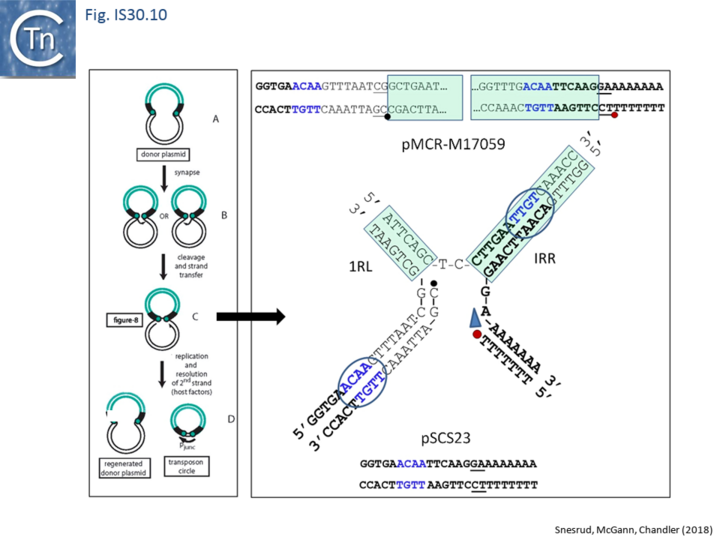
An observation which may be pertinent to this deletion phenomenon which leads to stabilisation of antibiotic resistance genes is the identification of a gene, iee (IS Enhanced Excision) originally implicated in enhanced excision of IS3 family member, IS629 [46]. The gene product, IEE, stimulates excision and adjacent deletions of certain types of IS. These are principally members of families, like IS30, which transpose using a copy-out-paste-in mechanism (see: IS3 family Excision). IEE contains potential primase and helicase motifs [46] (Fig. IS3.15) and although the authors suggest that it acts at the first strand transfer step which creates the bridged molecule, it seems possible that it acts during the copy-out replication step by slippage and realignment of the replication primer as proposed here (Fig. IS30.10). The gene is widespread in the bacterial kingdom [46] and probably carried by a mobile Integrative Conjugative Element. In this context, it seems possible that IEE may interfere with the normal transposition pathway by relaxing complementarity constraints of the replication fork.
Structural studies
There are no structural data available at present for any members of this family, although recent results obtained with an IS from another family, ISCth4 from the IS256 family, which also undergoes copy-out-paste-in transposition has provided some insights [47]. In accord with this type of mechanism, crystal structures of ISCth4 transposase bound to three different substrates show a transposase dimer bound asymmetrically to a single DNA substrate: a pre-reaction substrate with IRR together with its flanking DNA, a pre-cleaved complex in which the IRR flank had been removed and a strand transfer complex including an abutted IRR and IRL separated by a gapped 6 base pair linker (Fig. IS256.8). It is important to note that IS256 family transposases carry an alpha-helical insertion domain which separates the catalytic domain into two segments. This domain plays an important role in directing different DNA segments during the reaction. IS30 family transposases carry an uninterrupted catalytic domain without the alpha-helical insertion domain implying that the atomic details of the process will differ.
Bibliography
- ↑ Caspers P, Dalrymple B, Iida S, Arber W . IS30, a new insertion sequence of Escherichia coli K12. - Mol Gen Genet: 1984, 196(1);68-73 [PubMed:6090868] [DOI]
- ↑ 2.0 2.1 Arber W, Iida S, Jütte H, Caspers P, Meyer J, Hänni C . Rearrangements of genetic material in Escherichia coli as observed on the bacteriophage P1 plasmid. - Cold Spring Harb Symp Quant Biol: 1979, 43 Pt 2;1197-208 [PubMed:385224] [DOI]
- ↑ Ammendola S, Politi L, Scandurra R . Cloning and sequencing of ISC1041 from the archaeon Sulfolobus solfataricus MT-4, a new member of the IS30 family of insertion elements. - FEBS Lett: 1998 May 29, 428(3);217-23 [PubMed:9654137] [DOI]
- ↑ Smyth DS, Robinson DA . Integrative and sequence characteristics of a novel genetic element, ICE6013, in Staphylococcus aureus. - J Bacteriol: 2009 Oct, 191(19);5964-75 [PubMed:19648240] [DOI]
- ↑ 5.0 5.1 5.2 Dalrymple B . Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. - Mol Gen Genet: 1987 May, 207(2-3);413-20 [PubMed:3039299] [DOI]
- ↑ Rasmussen JL, Odelson DA, Macrina FL . Complete nucleotide sequence of insertion element IS4351 from Bacteroides fragilis. - J Bacteriol: 1987 Aug, 169(8);3573-80 [PubMed:3038844] [DOI]
- ↑ 7.0 7.1 Rudant E, Courvalin P, Lambert T . Characterization of IS18, an element capable of activating the silent aac(6')-Ij gene of Acinetobacter sp. 13 strain BM2716 by transposition. - Antimicrob Agents Chemother: 1998 Oct, 42(10);2759-61 [PubMed:9756793] [DOI]
- ↑ Sóki J, Eitel Z, Urbán E, Nagy E, ESCMID Study Group on Anaerobic Infections . Molecular analysis of the carbapenem and metronidazole resistance mechanisms of Bacteroides strains reported in a Europe-wide antibiotic resistance survey. - Int J Antimicrob Agents: 2013 Feb, 41(2);122-5 [PubMed:23158541] [DOI]
- ↑ 9.0 9.1 9.2 Stalder R, Caspers P, Olasz F, Arber W . The N-terminal domain of the insertion sequence 30 transposase interacts specifically with the terminal inverted repeats of the element. - J Biol Chem: 1990 Mar 5, 265(7);3757-62 [PubMed:2154486]
- ↑ 10.0 10.1 10.2 Nagy Z, Szabó M, Chandler M, Olasz F . Analysis of the N-terminal DNA binding domain of the IS30 transposase. - Mol Microbiol: 2004 Oct, 54(2);478-88 [PubMed:15469518] [DOI]
- ↑ 11.0 11.1 Calcutt MJ, Lavrrar JL, Wise KS . IS1630 of Mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion. - J Bacteriol: 1999 Dec, 181(24);7597-607 [PubMed:10601219] [DOI]
- ↑ Stalder R, Arber W . Characterization of in vitro constructed IS30-flanked transposons. - Gene: 1989, 76(2);187-93 [PubMed:2546856] [DOI]
- ↑ 13.0 13.1 13.2 Snesrud E, McGann P, Chandler M . The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: the Vehicle for Transferable Colistin Resistance. - mBio: 2018 Feb 13, 9(1); [PubMed:29440577] [DOI]
- ↑ 14.0 14.1 Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F . A Model for Transposition of the Colistin Resistance Gene mcr-1 by ISApl1. - Antimicrob Agents Chemother: 2016 Nov, 60(11);6973-6976 [PubMed:27620479] [DOI]
- ↑ 15.0 15.1 15.2 15.3 Li R, Chen K, Chan EW, Chen S . Characterization of the stability and dynamics of Tn6330 in an Escherichia coli strain by nanopore long reads. - J Antimicrob Chemother: 2019 Jul 1, 74(7);1807-1811 [PubMed:30927434] [DOI]
- ↑ 16.0 16.1 16.2 16.3 16.4 Olasz F, Stalder R, Arber W . Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements. - Mol Gen Genet: 1993 May, 239(1-2);177-87 [PubMed:8389976] [DOI]
- ↑ Naas T, Blot M, Fitch WM, Arber W . Insertion sequence-related genetic variation in resting Escherichia coli K-12. - Genetics: 1994 Mar, 136(3);721-30 [PubMed:7911771] [DOI]
- ↑ Dalrymple B, Caspers P, Arber W . Nucleotide sequence of the prokaryotic mobile genetic element IS30. - EMBO J: 1984 Sep, 3(9);2145-9 [PubMed:6092059] [DOI]
- ↑ 19.0 19.1 Dalrymple B, Arber W . Promotion of RNA transcription on the insertion element IS30 of E. coli K12. - EMBO J: 1985 Oct, 4(10);2687-93 [PubMed:2414100] [DOI]
- ↑ Arini A, Keller MP, Arber W . An antisense RNA in IS30 regulates the translational expression of the transposase. - Biol Chem: 1997 Dec, 378(12);1421-31 [PubMed:9461341] [DOI]
- ↑ Olasz F, Farkas T, Stalder R, Arber W . Mutations in the carboxy-terminal part of IS30 transposase affect the formation and dissolution of (IS30)2 dimer. - FEBS Lett: 1997 Aug 25, 413(3);453-61 [PubMed:9303555] [DOI]
- ↑ 22.0 22.1 Olasz F, Farkas T, Kiss J, Arini A, Arber W . Terminal inverted repeats of insertion sequence IS30 serve as targets for transposition. - J Bacteriol: 1997 Dec, 179(23);7551-8 [PubMed:9393723] [DOI]
- ↑ 23.0 23.1 23.2 23.3 23.4 Kiss J, Olasz F . Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. - Mol Microbiol: 1999 Oct, 34(1);37-52 [PubMed:10540284] [DOI]
- ↑ Kiss J, Szabó M, Olasz F . Site-specific recombination by the DDE family member mobile element IS30 transposase. - Proc Natl Acad Sci U S A: 2003 Dec 9, 100(25);15000-5 [PubMed:14665688] [DOI]
- ↑ Szeverényi I, Nagy Z, Farkas T, Olasz F, Kiss J . Detection and analysis of transpositionally active head-to-tail dimers in three additional Escherichia coli IS elements. - Microbiology (Reading): 2003 May, 149(Pt 5);1297-1310 [PubMed:12724391] [DOI]
- ↑ 26.0 26.1 26.2 Kiss J, Nagy Z, Tóth G, Kiss GB, Jakab J, Chandler M, Olasz F . Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis. - Mol Microbiol: 2007 Mar, 63(6);1731-47 [PubMed:17367392] [DOI]
- ↑ 27.0 27.1 Szabó M, Kiss J, Nagy Z, Chandler M, Olasz F . Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. - J Mol Biol: 2008 Jan 11, 375(2);337-52 [PubMed:18022196] [DOI]
- ↑ Hwa V, Shoemaker NB, Salyers AA . Direct repeats flanking the Bacteroides transposon Tn4351 are insertion sequence elements. - J Bacteriol: 1988 Jan, 170(1);449-51 [PubMed:2826402] [DOI]
- ↑ Brynestad S, Granum PE . Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. - FEMS Microbiol Lett: 1999 Jan 1, 170(1);281-6 [PubMed:9919679] [DOI]
- ↑ Reimmann C, Haas D . Mode of replicon fusion mediated by the duplicated insertion sequence IS21 in Escherichia coli. - Genetics: 1987 Apr, 115(4);619-25 [PubMed:3034717] [DOI]
- ↑ Turlan C, Ton-Hoang B, Chandler M . The role of tandem IS dimers in IS911 transposition. - Mol Microbiol: 2000 Mar, 35(6);1312-25 [PubMed:10760133] [DOI]
- ↑ Casadesús J, Naas T, Garzón A, Arini A, Torreblanca J, Arber W . Lack of hotspot targets: a constraint for IS30 transposition in Salmonella. - Gene: 1999 Sep 30, 238(1);231-9 [PubMed:10570999] [DOI]
- ↑ Olasz F, Kiss J, König P, Buzás Z, Stalder R, Arber W . Target specificity of insertion element IS30. - Mol Microbiol: 1998 May, 28(4);691-704 [PubMed:9643538] [DOI]
- ↑ Quast K, Bathe B, Pühler A, Kalinowski J . The Corynebacterium glutamicum insertion sequence ISCg2 prefers conserved target sequences located adjacent to genes involved in aspartate and glutamate metabolism. - Mol Gen Genet: 1999 Oct, 262(3);568-78 [PubMed:10589846] [DOI]
- ↑ Hu Y, Liu F, Lin IY, Gao GF, Zhu B . Dissemination of the mcr-1 colistin resistance gene. - Lancet Infect Dis: 2016 Feb, 16(2);146-7 [PubMed:26711359] [DOI]
- ↑ 36.0 36.1 36.2 Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J . Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. - Lancet Infect Dis: 2016 Feb, 16(2);161-8 [PubMed:26603172] [DOI]
- ↑ Zhi C, Lv L, Yu LF, Doi Y, Liu JH . Dissemination of the mcr-1 colistin resistance gene. - Lancet Infect Dis: 2016 Mar, 16(3);292-3 [PubMed:26973307] [DOI]
- ↑ Zurfluh K, Klumpp J, Nüesch-Inderbinen M, Stephan R . Full-Length Nucleotide Sequences of mcr-1-Harboring Plasmids Isolated from Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates of Different Origins. - Antimicrob Agents Chemother: 2016 Sep, 60(9);5589-91 [PubMed:27324774] [DOI]
- ↑ Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N . Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. - J Antimicrob Chemother: 2016 Aug, 71(8);2300-5 [PubMed:27090630] [DOI]
- ↑ Falgenhauer L, Waezsada SE, Gwozdzinski K, Ghosh H, Doijad S, Bunk B, Spröer C, Imirzalioglu C, Seifert H, Irrgang A, Fischer J, Guerra B, Käsbohrer A, Overmann J, Goesmann A, Chakraborty T . Chromosomal Locations of mcr-1 and bla CTX-M-15 in Fluoroquinolone-Resistant Escherichia coli ST410. - Emerg Infect Dis: 2016 Sep, 22(9);1689-91 [PubMed:27322919] [DOI]
- ↑ McGann P, Snesrud E, Maybank R, Corey B, Ong AC, Clifford R, Hinkle M, Whitman T, Lesho E, Schaecher KE . Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of mcr-1 in the United States. - Antimicrob Agents Chemother: 2016 Jul, 60(7);4420-1 [PubMed:27230792] [DOI]
- ↑ Tegetmeyer HE, Jones SC, Langford PR, Baltes N . ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. - Vet Microbiol: 2008 Apr 30, 128(3-4);342-53 [PubMed:18065168] [DOI]
- ↑ Schwarz S, Johnson AP . Transferable resistance to colistin: a new but old threat. - J Antimicrob Chemother: 2016 Aug, 71(8);2066-70 [PubMed:27342545] [DOI]
- ↑ 44.0 44.1 He YZ, Li XP, Miao YY, Lin J, Sun RY, Wang XP, Guo YY, Liao XP, Liu YH, Feng Y, Sun J . The ISApl1 (2) Dimer Circular Intermediate Participates in mcr-1 Transposition. - Front Microbiol: 2019, 10;15 [PubMed:30723461] [DOI]
- ↑ Szabó M, Kiss J, Kótány G, Olasz F . Importance of illegitimate recombination and transposition in IS30-associated excision events. - Plasmid: 1999 Nov, 42(3);192-209 [PubMed:10545262] [DOI]
- ↑ 46.0 46.1 46.2 Kusumoto M, Ooka T, Nishiya Y, Ogura Y, Saito T, Sekine Y, Iwata T, Akiba M, Hayashi T . Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. - Nat Commun: 2011 Jan 11, 2;152 [PubMed:21224843] [DOI]
- ↑ Kosek D, Hickman AB, Ghirlando R, He S, Dyda F . Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition. - EMBO J: 2021 Jan 4, 40(1);e105666 [PubMed:33006208] [DOI]