IS Families/IS481 family
Contents
- 1 Historical
- 2 Relationship to other IS
- 3 Transporter (t)IS derivatives
- 4 Use of IS481 for Diagnostic Purposes
- 5 Relationship to Eukaryotic IS
- 6 IS481 in the Evolution of Bordetella
- 7 Genomic sequences, IS expansion, genome streamlining and decay
- 8 Changes at the gene level
- 9 IS impact on gene expression
- 10 Acknowledgements
- 11 Bibliography
Historical
IS481 was first identified in the late 1980s [1][2] as a repeated sequence in the agent of whooping cough, Bordetella pertussis. It was estimated, from Southern blots of restriction digested B. pertusis genomic DNA, that IS481 was present in multiple copies of about 20, with some variation between strains. They were not observed as repeated sequences in the single strain of B. parapertussis or in B. bronchiseptica which were analyzed, although homology was observed as a single band in B. parapertussis strain BPAH1 genomic digests and two bands in digests of B. bronchiseptica strain BBRH1.
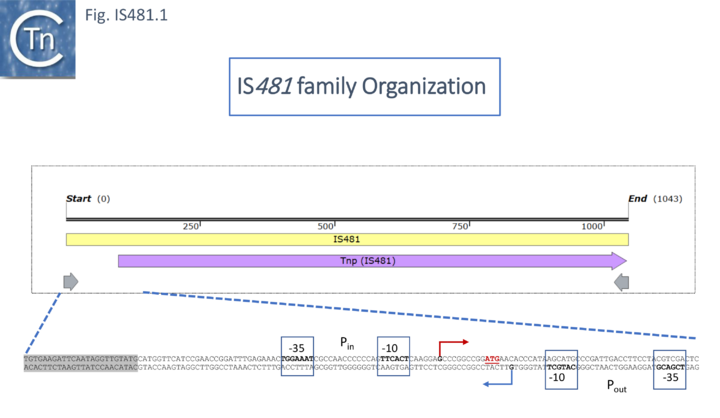
The DNA sequence [2][3][4], called IS481, was later identified as a 1052bp element with 28bp terminal inverted repeats and confirmed not tobe present in multiple copies in either theB. parapertussis or B. bronchiseptica genomes. This was later reassessed to be 1046bp in length (Fig. IS481.1; [5]) and to massively hybridize with genomic DNA of 100 B. pertusis clinical isolates. There were a number of small sequence variations observed in the results from different authors.
It was also observed to transpose. When the bvgA gene, a global virulence regulator, was present as both a plasmid and chromosomal copy, host growth was inhibited when the plasmid-borne gene carried a small deletion (a dominant negative mutation) [6]. Those derivatives in which growth inhibition was relieved had acquired IS481 insertions into the plasmid-borne gene or IS481/IS1002 insertions into the chromosomal gene and to generate 6bp target DR [7].
Relationship to other IS
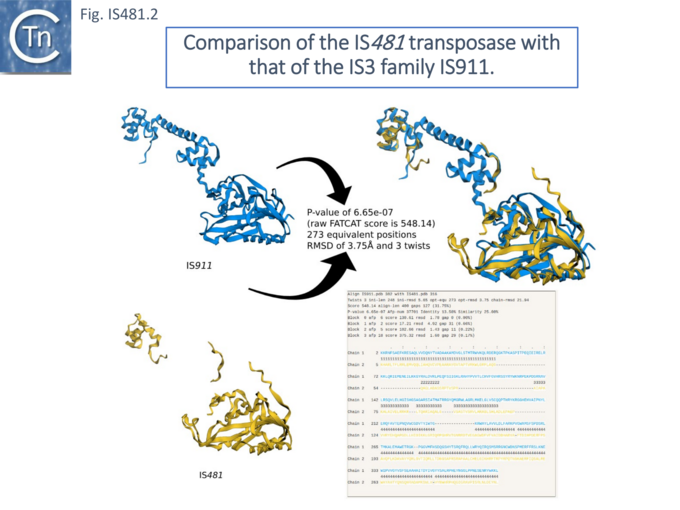
Initially, IS481 appeared to be an IS3 family derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in 5’ TGT 3’) are similar to those of IS3 family members. Although, at that time IS481 had not been demonstrated to transpose, their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggested that these represent a distinct transpositionally active family. Different members generate DR of between 4 and 15 bp. Moreover, certain members (e.g. ISSav7) insert specifically into the tetranucleotide CTAG which becomes the flanking DR and provides the UAG termination codon for the Tpase. In contrast to the vast majority of IS3 family members, the IS481 Tpase is not produced by frameshifting. There is no evidence for a leucine zipper as in IS3. This can be clearly seen from the alignment and remarkable predicted structural similarities in the C-terminal catalytic domain and small N-terminal domain (Fig. IS481.2).
They also show a conserved 5’TG 3’ tip to the IR which is typical of this and a number of other types of prokaryotic mobile elements.
IS481 is also distantly related to the IS1202 family [9][10].
Transporter (t)IS derivatives
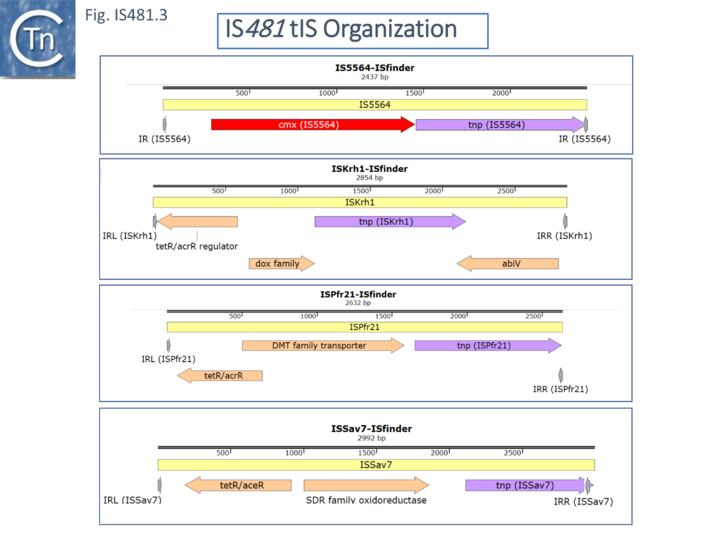
Some members carry passenger genes (Fig. IS481.3) including antibiotic resistance (CmR, cmx, for IS5564 and the closely related ISCgl1), or potential transcriptional regulators among other open reading frames (ISKrh1, ISPfr21- which contains a termination codon within the transposase, and ISSav7).
Use of IS481 for Diagnostic Purposes
There are PCR diagnostic clinical protocols for Bordetella based on IS481 [12][13][14] although several instances of false positive results have been reported.
Relationship to Eukaryotic IS
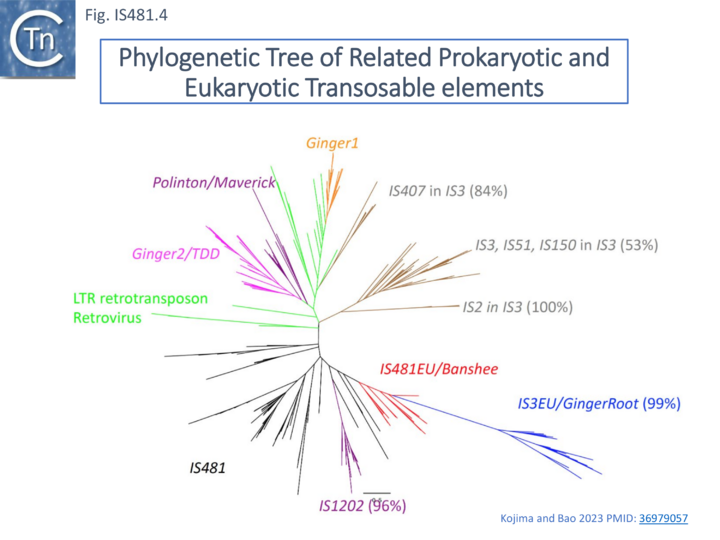
IS481 family IS are distantly related to the eukaryotic Banshee transposon which at present is restricted to the anaerobic flagellated protozoan Trichomonas vaginalis [15][16]. They share the highly conserved Pfam integrase core domain identified in the IS3 family and retroviruses [17][18]. Kojima and Bao [16], have identified a number of related IS from Trichomonas vaginalis which they have called IS481EU.
Interestingly, these were not well separated from a group of IS3 family-related eukaryotic sequences they called IS3EU/GingerRoot (similarly to their prokaryotic relatives) (Fig. IS481.4). It would be interesting to determine whether IS481EU transpose using a dsDNA circular intermediate as do IS3 family members.
IS481 in the Evolution of Bordetella
IS481 itself has played a fundamental role in the evolution of the genomes of the Bordetellae. The genus Bordetella includes 16 named species (see Wigland et al [19]). Three of these, B. bronchiseptica, B. parapertussis and B. pertussis are closely related and colonize the mammalian respiratory tract. Of the three, B. bronchiseptica is thought to be ancestral to B. parapertussis and B. pertussis although B. parapertussis is more closely related to B. bronchiseptica than to B. pertussis [20]. While B. bronchiseptica can infect a number of mammals, B. pertussis is a strict human pathogen and B. parapertussis infects both humans and sheep.
More detailed analysis revealed a relative of IS481, IS1002, with identical IR but only 61.5% sequence identity [21] was present in B. pertussis and B. parapertussis strains from humans but absent in B. parapertussis isolated from sheep, suggesting that little or no transmission has occurred between sheep and humans.
It was postulated that, once in the human host, B. parapertussis probably acquired IS1002 from B. pertussis. In contrast to human B. parapertussis, isolates, B. pertussis strains produced polymorphic IS1002-related DNA fingerprint patterns [21].
Genomic sequences, IS expansion, genome streamlining and decay
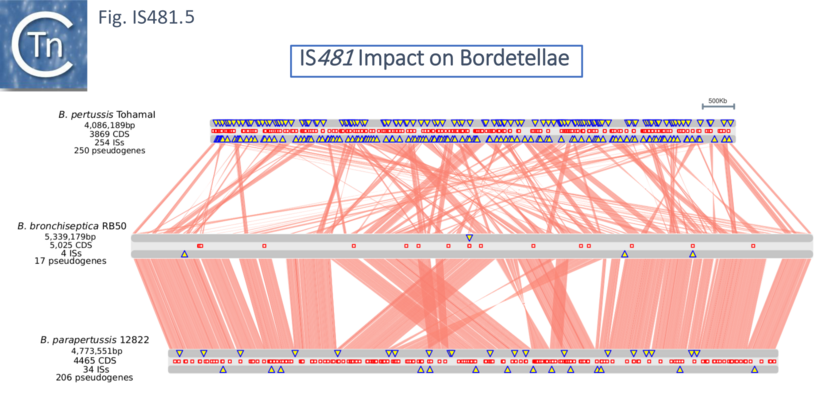
A remarkable result came from a comparison of the whole genome sequences of the three Bordatellae: B. bronchiseptica RB50; B. parapertussis 12822); and B. pertussis Tohama I [22] (Fig. IS481.5) showing a large expansion in one particular IS, IS481 accompanied by generation of an extensive collection of pseudogenes.
This analysis confirmed that B. parapertussis and B. pertussis are independent derivatives of B. bronchiseptica-like ancestors [20]. Indeed, it has even been proposed that B. pertussis has evolved from a human-associated B. bronchiseptica lineage [23]. Moreover, the process was driven by prophage loss from B. bronchiseptica and a massive amplification of IS481/IS1002 copies (261 in B. pertussis and 112 in B. parapertussis; [24]) accompanied by significant genome reduction and extensive gene inactivation and loss (Fig. IS481.5) and in the two host-restricted species. Although there are a number of collinear regions between B. bronchiseptica and B. parapertussis, most of the rearrangements involve either IS1001 or the IS481-related IS1002.
The rearrangements and deletions in B. pertussis were similar to those in B. parapertussis but more extreme. Short, highly conserved, segments were observed to be fragmented into ~150 individual rearrangements, with the vast majority (88%) bounded primarily by copies of IS481. Thus, the authors point out, adaptation to specific hosts results in loss rather than gain of function. A study which also included B. holmesii found that inversions in the genomes of B. holmesii, B. pertussis, and B. parapertussis were generally flanked by the multicopy IS which occur in their genomes [19].
Changes at the gene level
There is little net gain of genes in the host restrictive species: very few genes were present in B. pertussis and B. parapertussis but missing in B. bronchiseptica RB50. On the other hand, B. bronchiseptica had over 600 genes not present in either of the other two species: over 1,000 were shared only with B. parapertussis while just over 100 were shared only with B. pertussis. Moreover, IS expansion was accompanied by a large increase in the number of pseudogenes (Fig. IS481.5).
IS impact on gene expression
It was suggested that IS481 might play an important functional role for B. pertussis in facilitating its adaptation to new environmental conditions by controlling characters such as phase variation and production of virulence determinants. In this light, IS481 was shown to drive expression of the catalase gene katA in strain BP504 [25] and the type III cytotoxic effector gene bteA, responsible for host cell death in Bordetella bronchiseptica infections in the vaccine-derived strain BP155 [26].
A more recent study [27] addressed the global impact of IS on the the B. pertussis transcriptome due to the presence of IS-associated promoters (Fig. IS481.1 bottom) or the formation of hybrid promoters on insertion. This study revealed that that most IS481 elements impact the transcriptional level of their flanking genes and can result in an uncharacterized regulation in a strain-specific manner due to significant strain to strain IS copy number variation and variations in their genomic context. Using the B. pertussis Tohama I strain the authors observed, from both RNA-seq and an approach to map Transcriptional Start Sites (TSS), that a significant number of IS generated transcripts into neighboring genes from a promoter at the left end (Fig. IS481.1; Fig.IS481.6) which they called Pout with reference to a similar promoter in IS10 which produces an RNA involved in regulation of transposase expression (IS10 [28][29][30][31] and in other IS (e.g. IS200 [32] and ISPpu9 [33] ).
Transcripts were identified extending into the flanking gene from Pout both in the “sense” orientation, potentially activating its transcription (70 IS: 66 copies of IS481 and 2 of IS1002 as well as 2 IS110 family IS1663), and in the antisense direction, potentially inhibiting expression (150 IS: 139 IS481 copies, 2 of IS1002 and 9 IS110 family IS1663).
Many cases were observed where Pin-driven transposase transcription escapes into a downstream gene from the right end (97 IS481, 1 IS1002 and 3 IS1663), in the same (46 times) or opposite (38 times) orientation.
Perhaps rather surprisingly, in view of their abundance in other systems [34][35], only 6 IS481 transcripts originated from a hybrid promoter in which the inserted IS provides a -35 promoter element to a endogenous -10 element.
The effects of these IS-associated transcripts on virulence, pathogenicity and persistence is gradually being assessed in detail. Pout had been identified by its ability to drive a neighboring katA gene in B. pertussis BP504 [25] and a bteA gene whose product is cytotoxic, in the vaccine-derived BP155 strain [26]. Interestingly the IS481 insertion occurred in B. pertussis clinical strains and the protein was found at higher levels in B. pertussis nonvaccine-type strains than in vaccine-type strains. The type-dependent expression was due to an insertion of IS481.
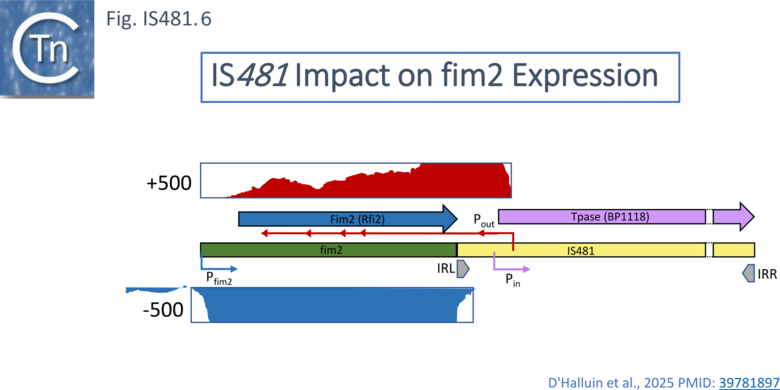
In the B. pertussisTohama I strain study, one of the IS481 regulatory RNAs (asRNAs) transcript transcribed from Pout was observed to impinge antisense to and attenuate fim2 gene expression. fim2 encodes serotype 2 fimbriae virulence factors [8] and was chosen for further study. This mutation, Rfi2, was found to be present only in B. pertussis and results in decreased cytotoxicity. Moreover, it was suggested that, in view of its immunogenicity, reduced Fim2 expression “contributes to immune evasion”. RNA seq confirmed a substantial level of transcription from Pout into the fim2 gene in the antisense-orientation and also revealed the presence of a number of prematurely truncated transcripts (Fig.IS481.6).
Acknowledgements
We would like to than Kenji Kojima for kindly supplying Figure 4.
Bibliography
- ↑ McPheat WL, McNally T . Distribution of a repeated DNA sequence in the genus "Bordetella". - Ann Sclavo Collana Monogr: 1986, 3(1-2);313-8 [PubMed:3426877]
- ↑ 2.0 2.1 McPheat WL, McNally T . Isolation of a repeated DNA sequence from Bordetella pertussis. - J Gen Microbiol: 1987 Feb, 133(2);323-30 [PubMed:2888834] [DOI]
- ↑ 3.0 3.1 McLafferty MA, Harcus DR, Hewlett EL . Nucleotide sequence and characterization of a repetitive DNA element from the genome of Bordetella pertussis with characteristics of an insertion sequence. - J Gen Microbiol: 1988 Aug, 134(8);2297-306 [PubMed:2908119] [DOI]
- ↑ McPheat WL, Hanson JH, Livey I, Robertson JS . Analysis of the chromosomal location of two copies of a Bordetella pertussis insertion sequence. - Mol Microbiol: 1989 Jul, 3(7);985-9 [PubMed:2552259] [DOI]
- ↑ 5.0 5.1 Glare EM, Paton JC, Premier RR, Lawrence AJ, Nisbet IT . Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. - J Clin Microbiol: 1990 Sep, 28(9);1982-7 [PubMed:2229381] [DOI]
- ↑ Stibitz S . Mutations affecting the alpha subunit of Bordetella pertussis RNA polymerase suppress growth inhibition conferred by short C-terminal deletions of the response regulator BvgA. - J Bacteriol: 1998 May, 180(9);2484-92 [PubMed:9573202] [DOI]
- ↑ Stibitz S . IS481 and IS1002 of Bordetella pertussis create a 6-base-pair duplication upon insertion at a consensus target site. - J Bacteriol: 1998 Sep, 180(18);4963-6 [PubMed:9733704] [DOI]
- ↑ 8.0 8.1 8.2 D'Halluin A, Petráčková D, Čurnová I, Držmíšek J, Čapek J, Bouquet P, Henin L, Antoine R, Coutte L, Locht C, Večerek B, Hot D . An IS element-driven antisense RNA attenuates the expression of serotype 2 fimbriae and the cytotoxicity of Bordetella pertussis. - Emerg Microbes Infect: 2025 Dec, 14(1);2451718 [PubMed:39781897] [DOI]
- ↑ Siguier P, Rousseau P, Cornet F, Chandler M . A subclass of the IS1202 family of bacterial insertion sequences targets XerCD recombination sites. - Plasmid: 2023 Jun 9, 127;102696 [PubMed:37302728] [DOI]
- ↑ Harmer CJ, Pong CH, Hall RM . Insertion sequences related to ISAjo2 target pdif and dif sites and belong to a new IS family, the IS1202 family. - Microb Genom: 2023 Mar, 9(3); [PubMed:36880881] [DOI]
- ↑ Ye Y, Godzik A . FATCAT: a web server for flexible structure comparison and structure similarity searching. - Nucleic Acids Res: 2004 Jul 1, 32(Web Server issue);W582-5 [PubMed:15215455] [DOI]
- ↑ van der Zee A, Agterberg C, Peeters M, Schellekens J, Mooi FR . Polymerase chain reaction assay for pertussis: simultaneous detection and discrimination of Bordetella pertussis and Bordetella parapertussis. - J Clin Microbiol: 1993 Aug, 31(8);2134-40 [PubMed:8370741] [DOI]
- ↑ Peng Y, Williams MM, Xiaoli L, Simon A, Fueston H, Tondella ML, Weigand MR . Strengthening Bordetella pertussis genomic surveillance by direct sequencing of residual positive specimens. - J Clin Microbiol: 2024 Apr 10, 62(4);e0165323 [PubMed:38445858] [DOI]
- ↑ Reischl U, Lehn N, Sanden GN, Loeffelholz MJ . Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. - J Clin Microbiol: 2001 May, 39(5);1963-6 [PubMed:11326023] [DOI]
- ↑ Feschotte C, Pritham EJ . DNA transposons and the evolution of eukaryotic genomes. - Annu Rev Genet: 2007, 41;331-68 [PubMed:18076328] [DOI]
- ↑ 16.0 16.1 16.2 Kojima KK, Bao W . IS481EU Shows a New Connection between Eukaryotic and Prokaryotic DNA Transposons. - Biology (Basel): 2023 Feb 25, 12(3); [PubMed:36979057] [DOI]
- ↑
Fayet O, Ramond P, Polard P, Prère MF, Chandler M . Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? - Mol Microbiol: 1990 Oct, 4(10);1771-7 [PubMed:1963920]
[DOI]
- ↑ Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM . Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. - Mol Cell Biol: 1992 May, 12(5);2331-8 [PubMed:1314954] [DOI]
- ↑ 19.0 19.1 Weigand MR, Peng Y, Batra D, Burroughs M, Davis JK, Knipe K, Loparev VN, Johnson T, Juieng P, Rowe LA, Sheth M, Tang K, Unoarumhi Y, Williams MM, Tondella ML . Conserved Patterns of Symmetric Inversion in the Genome Evolution of Bordetella Respiratory Pathogens. - mSystems: 2019 Nov 19, 4(6); [PubMed:31744907] [DOI]
- ↑ 20.0 20.1 Musser JM, Hewlett EL, Peppler MS, Selander RK . Genetic diversity and relationships in populations of Bordetella spp. - J Bacteriol: 1986 Apr, 166(1);230-7 [PubMed:3957867] [DOI]
- ↑ 21.0 21.1 van der Zee A, Groenendijk H, Peeters M, Mooi FR . The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. - Int J Syst Bacteriol: 1996 Jul, 46(3);640-7 [PubMed:8782670] [DOI]
- ↑ Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ . Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. - Nat Genet: 2003 Sep, 35(1);32-40 [PubMed:12910271] [DOI]
- ↑ Diavatopoulos DA, Cummings CA, Schouls LM, Brinig MM, Relman DA, Mooi FR . Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. - PLoS Pathog: 2005 Dec, 1(4);e45 [PubMed:16389302] [DOI]
- ↑ Preston A, Parkhill J, Maskell DJ . The bordetellae: lessons from genomics. - Nat Rev Microbiol: 2004 May, 2(5);379-90 [PubMed:15100691] [DOI]
- ↑ 25.0 25.1 DeShazer D, Wood GE, Friedman RL . Molecular characterization of catalase from Bordetella pertussis: identification of the katA promoter in an upstream insertion sequence. - Mol Microbiol: 1994 Oct, 14(1);123-30 [PubMed:7830550] [DOI]
- ↑ 26.0 26.1 Han HJ, Kuwae A, Abe A, Arakawa Y, Kamachi K . Differential expression of type III effector BteA protein due to IS481 insertion in Bordetella pertussis. - PLoS One: 2011 Mar 10, 6(3);e17797 [PubMed:21423776] [DOI]
- ↑ Amman F, D'Halluin A, Antoine R, Huot L, Bibova I, Keidel K, Slupek S, Bouquet P, Coutte L, Caboche S, Locht C, Vecerek B, Hot D . Primary transcriptome analysis reveals importance of IS elements for the shaping of the transcriptional landscape of Bordetella pertussis. - RNA Biol: 2018, 15(7);967-975 [PubMed:29683387] [DOI]
- ↑ Simons RW, Hoopes BC, McClure WR, Kleckner N . Three promoters near the termini of IS10: pIN, pOUT, and pIII. - Cell: 1983 Sep, 34(2);673-82 [PubMed:6311437] [DOI]
- ↑
Simons RW, Kleckner N . Translational control of IS10 transposition. - Cell: 1983 Sep, 34(2);683-91 [PubMed:6311438]
[DOI]
- ↑ Case CC, Roels SM, González JE, Simons EL, Simons RW . Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. - Gene: 1988 Dec 10, 72(1-2);219-36 [PubMed:2468561] [DOI]
- ↑ Ma C, Simons RW . The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. - EMBO J: 1990 Apr, 9(4);1267-74 [PubMed:1691097] [DOI]
- ↑ Ellis MJ, Haniford DB . Riboregulation of bacterial and archaeal transposition. - Wiley Interdiscip Rev RNA: 2016 May, 7(3);382-98 [PubMed:26846462] [DOI]
- ↑ Gómez-García G, Ruiz-Enamorado A, Yuste L, Rojo F, Moreno R . Expression of the ISPpu9 transposase of Pseudomonas putida KT2440 is regulated by two small RNAs and the secondary structure of the mRNA 5'-untranslated region. - Nucleic Acids Res: 2021 Sep 20, 49(16);9211-9228 [PubMed:34379788] [DOI]
- ↑ Vandecraen J, Chandler M, Aertsen A, Van Houdt R . The impact of insertion sequences on bacterial genome plasticity and adaptability. - Crit Rev Microbiol: 2017 Nov, 43(6);709-730 [PubMed:28407717] [DOI]
- ↑ Siguier P, Gourbeyre E, Chandler M . Bacterial insertion sequences: their genomic impact and diversity. - FEMS Microbiol Rev: 2014 Sep, 38(5);865-91 [PubMed:24499397] [DOI]