IS Families/IS481 family
Contents
- 1 Historical
- 2 Relationship to other IS.
- 3 Transporter (t)IS derivatives
- 4 Use of IS481 for Diagnostic Purposes
- 5 Relationship to Eukaryotic IS
- 6 IS481 in the Evolution of Bordetella
- 7 Genomic sequences, IS expansion, genome streamlining and decay
- 8 Changes at the gene level
- 9 General
- 10 Bibliography
Historical
IS481 was first identified in the late 1980s [1][2] as a repeated sequence in the agent of whooping cough, Bordetella pertussis. It was estimated, from Southern blots of restriction digested B. pertusis genomic DNA, that IS481 was present in multiple copies of about 20, with some variation between strains. They were not observed as repeated sequences in the single strain of B. parapertussis or in B. bronchiseptica which were analyzed, although homology was observed as a single band in B. parapertussis strain BPAH1 genomic digests and two bands in digests of B. bronchiseptica strain BBRH1.
The DNA sequence [2][3][4] later identified IS481 as a 1052bp element with 28bp terminal inverted repeats and confirmed that it was not present in multiple copies in either theB. parapertussis or B. bronchiseptica genomes. This was later reassessed to be 1046bp in length (Fig. IS481.1; [5]) and to massively hybridize with pertussis genomic DNA of 100 B. pertusis clinical isolates. There were a number of small sequence variations observed in the results from different authors.
It was also observed to transpose. When the bvgA gene, a global virulence regulator, was present as both a plasmid and chromosomal copy, host growth was inhibited when the plasmid-borne gene carries a small deletion [6]. Those derivatives in which growth inhibition was relieved had acquired IS481 insertions into the plasmid-borne gene or IS481/IS1002 insertions into the chromosomal gene and to generate 6bp target DR [7].
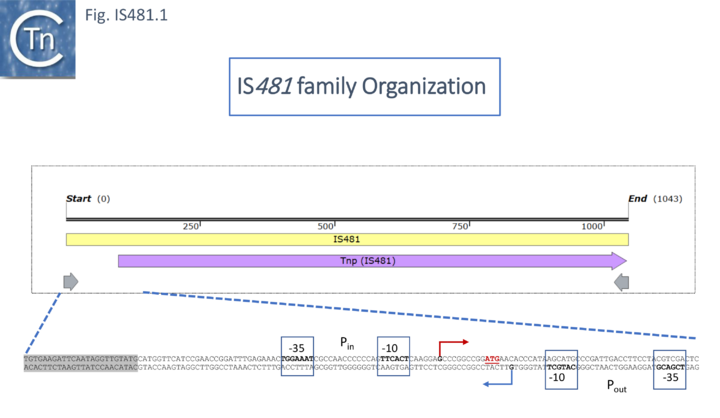
Relationship to other IS.
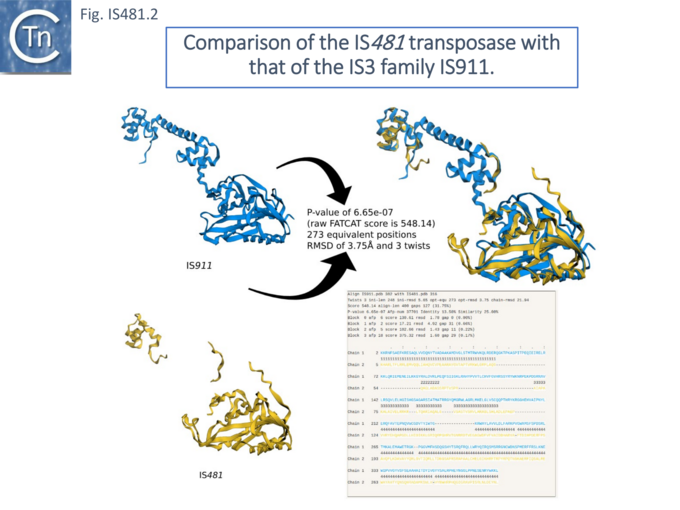
Initially, IS481 appeared to be an IS3 family derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in 5’ TGT 3’) are similar to those of IS3 family members. Although, at that time IS481 had not been demonstrated to transpose, their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggested that these represent a distinct transpositionally active family. Different members generate DR of between 4 and 15 bp. Moreover, certain members (e.g. ISSav7) insert specifically into the tetranucleotide CTAG which becomes the flanking DR and provides the UAG termination codon for the Tpase. In contrast to the vast majority of IS3 family members, the IS481 Tpase is not produced by frameshifting. There is no evidence for a leucine zipper as in IS3. This can be clearly seen from the alignment and remarkable predicted structural similarities in the C-terminal catalytic domain and small N-terminal domain (Fig. IS481.2).
They also show a conserved 5’TG 3’ tip to the IR which is typical of this and a number of other types of prokaryotic mobile elements.
IS481 is also distantly related to the IS1202 family [9][10].
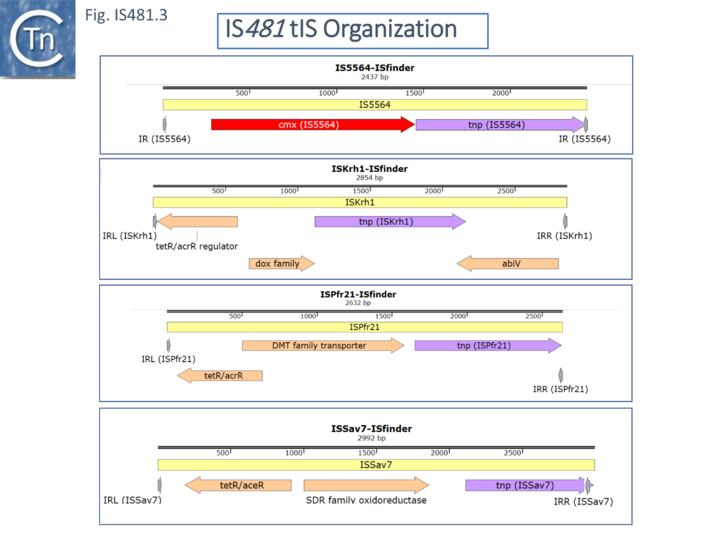
Transporter (t)IS derivatives
Some members carry passenger genes (Fig. IS481.3) including antibiotic resistance (CmR, cmx, for IS5564 and the closely related ISCgl1), or potential transcriptional regulators among other open reading frames (ISKrh1, ISPfr21- which contains a termination codon within the transposase, and ISSav7).
Use of IS481 for Diagnostic Purposes
There are PCR diagnostic clinical protocols for Bordetella based on IS481 [12][13][14] although several instances of false positive results have been reported.
Relationship to Eukaryotic IS
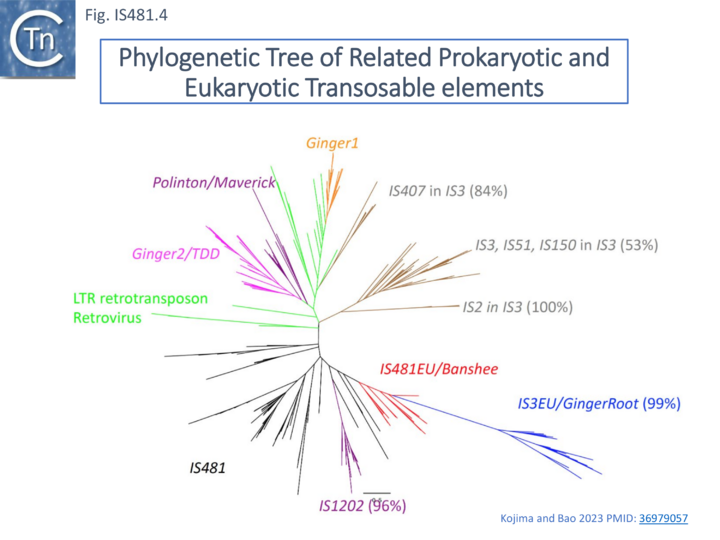
IS481 family IS are distantly related to the eukaryotic Banshee transposon which at present is restricted to the anaerobic flagellated protozoan Trichomonas vaginalis [15][16]. They share the highly conserved Pfam integrase core domain identified in the IS3 family and retroviruses [17][18]. Kojima and Bao [16], have identified a number of related IS from Trichomonas vaginalis which they have called IS481EU.
Interestingly, these were not well separated from a group of IS3 family-related eukaryotic sequences they called IS3EU/GingerRoot (similarly to the prokaryotic relatives) (Fig. IS481.4). It would be interesting to determine whether IS481EU transpose using a dsDNA circular intermediate as do IS3 family members.
IS481 in the Evolution of Bordetella
IS481 itself has played a fundamental role in the evolution of the genomes of the Bordetellae. The genus Bordetella includes 16 named species (see Wigland et al [19]). Three of these, B. bronchiseptica, B. parapertussis and B. pertussis are closely related and colonize the mammalian respiratory tract. Of the three, B. bronchiseptica is thought to be ancestral to B. parapertussis and B. pertussis although B. parapertussis is more closely related to B. bronchiseptica than to B. pertussis [20]. While B. bronchiseptica can infect a number of mammals, B. pertussis is a strict human pathogen and B. parapertussis infects both humans and sheep.
More detailed analysis revealed a relative of IS481, IS1002, with identical IR but only 61.5% sequence identity [21] was present in B. pertussis and B. parapertussis strains from humans but absent in B. parapertussis isolated from sheep, suggesting that little or no transmission has occurred between sheep and humans.
It was postulated that, once in the human host, B. parapertussis probably acquired IS1002 from B. pertussis. In contrast to human B. parapertussis, isolates, B. pertussis strains produced polymorphic IS1002-related DNA fingerprint patterns [21].
Genomic sequences, IS expansion, genome streamlining and decay
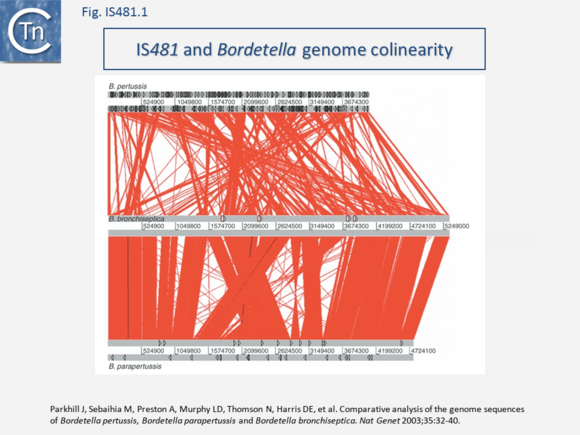
A remarkable result came from a comparison of the whole genome sequences of the three Bordatellae: B. bronchiseptica RB50; B. parapertussis 12822); and B. pertussis Tohama I (Parkhill et al., 2003 PMID: 12910271)(Fig. IS481.4) showing a large expansion in one particular IS, IS481 accompanied by generation of an extensive collection of pseudogenes. (see……..????)
This analysis confirmed that B. parapertussis and B. pertussis are independent derivatives of B. bronchiseptica-like ancestors (Musser et al., 1986 PMID: 3957867). Indeed, it has even been proposed that B. pertussis has evolved from a human-associated B. bronchiseptica lineage (Diavatopoulos et al, 2005 PMID: 16389302). Moreover, the process was driven by prophage loss from B. bronchiseptica and a massive amplification of IS481/IS1002 copies (261 in B. pertussis and 112 in B. parapertussis; Preston et al 2004 PMID: 15100691) accompanied by significant genome reduction and extensive gene inactivation and loss (Fig. IS481.XXX) and in the two host-restricted species. Although there are a number of collinear regions between B. bronchiseptica and B. parapertussis, most of the rearrangements involve either IS1001 or the IS481-related IS1002.
The rearrangements and deletions in B. pertussis were similar to those in B. parapertussis but more extreme. Short, highly conserved, segments were observed to be fragmented into ~150 individual rearrangements, with the vast majority (88%) bounded primarily by copies of IS481. Thus, the authors point out, adaptation to specific hosts results in loss rather than gain of function. A study which also included B. holmesii found that inversions in the genomes of B. holmesii, B. pertussis, and B. parapertussis were generally flanked by the multicopy IS which occur in their genomes (Wigland et al PMID: 31744907).
Changes at the gene level
There is little net gain of genes in the host restrictive species: very few genes were present in B. pertussis and B. parapertussis but missing in B. bronchiseptica RB50. On the other hand, B. bronchiseptica had over 600 genes not present in either of the other two species: over 1,000 were shared only with B. parapertussis while just over 100 were shared only with B. pertussis. Moreover, IS expansion was accompanied by a large increase in the number of pseudogenes (Fig. IS481.3).
General
Initially, IS481 appeared to be an IS3 family derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in 5’ TGT 3’) are similar to those of IS3 family members. Their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggests that these represent a distinct transpositionally active family.
Different members generate DR of between 4 and 15 bp. Moreover, certain members (e.g. ISSav7) insert specifically into the tetranucleotide CTAG which becomes the flanking DR and provides the UAG termination codon for the Tpase. In contrast to the vast majority of IS3 family members, the IS481 Tpase is not produced by frameshifting. There is no evidence for a leucine zipper as in IS3.
Some members include passenger genes including antibiotic resistance (Chloramphenicol - CmR gene for IS5564 and ISCgl1), or potential transcriptional regulators (ISKrh1, ISPfr21, ISSav7). IS481 itself has played a fundamental role in the evolution of the genomes of the Bordetellae where, in B. pertusis it has undergone extensive amplification to several hundred copies with accompanying genome decay[22][23] (Fig.IS481.1),
These IS are distantly related to the eukaryotic Banshee transposon which at present is restricted to the anaerobic flagellated protozoan Trichomonas vaginalis (Pritham per. comm.)[24]. They share the highly conserved Pfam integrase core domain identified initially in the IS3 family and retroviruses[25][26]. They also show a conserved 5’ TG 3’ tip to the IR which is typical of this and other types of mobile elements. It would be interesting to determine whether Banshee transposes using a dsDNA circular intermediate as do IS3 family members.
IS1202 group (ISNCY)
A small group including IS1202[27], which had been included in the ISNCY (Not Classified Yet) group appears distantly related to IS481. Members are between 1400 and 1700 bp (except for ISKpn21 which includes a passenger gene annotated as "hypothetical protein”) with a Tpase orf of between 400 and 500 aa in a single reading frame. Their IR begin with TGT as do those of the IS3 and IS481 families. They generate DR of between 5 and, unusually, 27 bp.
They appear to have similarities at the level of their Tpases particularly in their DDE domains (e.g. IS1202 is 39% aa similar to ISPfr5 of the IS481 family). Furthermore, they include glutamine (Q) seven residues C-terminal to the conserved E instead of the characteristic K/R. Identification of additional IS will be necessary to clearly define this group.
Bibliography
- ↑ <pubmed>3426877</pubmed>
- ↑ 2.0 2.1 <pubmed>2888834</pubmed>
- ↑ 3.0 3.1 <pubmed>2908119</pubmed>
- ↑ <pubmed>2552259</pubmed>
- ↑ 5.0 5.1 <pubmed>2229381</pubmed>
- ↑ <pubmed>9573202</pubmed>
- ↑ <pubmed>9733704</pubmed>
- ↑ <pubmed>39781897</pubmed>
- ↑ <pubmed>37302728</pubmed>
- ↑ <pubmed>36880881</pubmed>
- ↑ <pubmed>15215455</pubmed>
- ↑ <pubmed>8370741</pubmed>
- ↑ <pubmed>38445858</pubmed>
- ↑ <pubmed>11326023</pubmed>
- ↑ <pubmed>18076328</pubmed>
- ↑ 16.0 16.1 16.2 <pubmed>36979057</pubmed>
- ↑ <pubmed>1963920</pubmed>
- ↑ <pubmed>1314954</pubmed>
- ↑ <pubmed>31744907</pubmed>
- ↑ <pubmed>3957867</pubmed>
- ↑ 21.0 21.1 <pubmed>8782670</pubmed>
- ↑ Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, Harris DE, Holden MT, Churcher CM, Bentley SD, Mungall KL, Cerdeño-Tárraga AM, Temple L, James K, Harris B, Quail MA, Achtman M, Atkin R, Baker S, Basham D, Bason N, Cherevach I, Chillingworth T, Collins M, Cronin A, Davis P, Doggett J, Feltwell T, Goble A, Hamlin N, Hauser H, Holroyd S, Jagels K, Leather S, Moule S, Norberczak H, O'Neil S, Ormond D, Price C, Rabbinowitsch E, Rutter S, Sanders M, Saunders D, Seeger K, Sharp S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Unwin L, Whitehead S, Barrell BG, Maskell DJ . Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. - Nat Genet: 2003 Sep, 35(1);32-40 [PubMed:12910271] [DOI]
- ↑ Preston A, Parkhill J, Maskell DJ . The bordetellae: lessons from genomics. - Nat Rev Microbiol: 2004 May, 2(5);379-90 [PubMed:15100691] [DOI]
- ↑ Feschotte C, Pritham EJ . DNA transposons and the evolution of eukaryotic genomes. - Annu Rev Genet: 2007, 41;331-68 [PubMed:18076328] [DOI]
- ↑ Fayet O, Ramond P, Polard P, Prère MF, Chandler M . Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? - Mol Microbiol: 1990 Oct, 4(10);1771-7 [PubMed:1963920] [DOI]
- ↑ Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM . Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. - Mol Cell Biol: 1992 May, 12(5);2331-8 [PubMed:1314954] [DOI]
- ↑ Morona JK, Guidolin A, Morona R, Hansman D, Paton JC . Isolation, characterization, and nucleotide sequence of IS1202, an insertion sequence of Streptococcus pneumoniae. - J Bacteriol: 1994 Jul, 176(14);4437-43 [PubMed:8021229] [DOI]