Difference between revisions of "IS Families/ISPa17 family"
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
====General==== | ====General==== | ||
| − | In contrast to other IS families, little attention has been paid to and element called [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. It was first identified as an autonomous unit in the sequence of the IncP-6 plasmid Rms149 <ref name=":0" | + | In contrast to other IS families, little attention has been paid to and element called [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. It was first identified as an autonomous unit in the sequence of the IncP-6 plasmid Rms149 <ref name=":0"><pubmed>15995187</pubmed></ref> along with a number of other potential TE and was shown as an element including 3 orfs. Haines et al note that it had been observed previously in the ''[[wikipedia:Pseudomonas_aeruginosa|Pseudomonas aeruginosa]]'' plasmid pKLC102 as three ''orfs'' located downstream from an integron Int1 gene but not identified as an autonomous IS presumably because it was not flanked by a direct target repeat and did not exhibit a recognizable transposase <ref><pubmed>14702321</pubmed></ref>. |
| − | The Rms149 copy was flanked by a 5 bp direct repeat (CGCGG) and was bordered by 25bp terminal inverted repeats ([[:File:ISPa17-fig1.png|Fig. ISPa17.1A]])<ref name=":0" />. It was located upstream from a [[Transposons families/Tn3 family|Tn''3''-family]] transposase. Further analysis (Chandler unpublished observation) revealed 4 orfs, two of which constitute a toxin-antitoxin system related to [[wikipedia:VapBC|''vapBC'' gene]] which is also present in the [[Transposons families/Tn3 family|Tn''3'' family transposon]] [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'']. A third ''orf'' was tentatively identified as a recombinase while a function for the fourth could not be identified. | + | The Rms149 copy was flanked by a 5 bp direct repeat (CGCGG) and was bordered by 25bp terminal inverted repeats ([[:File:ISPa17-fig1.png|Fig. ISPa17.1A]])<ref name=":0" />. It was located upstream from a [[Transposons families/Tn3 family|Tn''3''-family]] transposase. Further analysis (Chandler unpublished observation) revealed 4 orfs, two of which constitute a [[wikipedia:Toxin-antitoxin_system|toxin-antitoxin system]] related to [[wikipedia:VapBC|''vapBC'' gene]] which is also present in the [[Transposons families/Tn3 family|Tn''3'' family transposon]] [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'']. A third ''orf'' was tentatively identified as a recombinase while a function for the fourth could not be identified. |
| − | [[File:ISPa17-fig1.png|center|thumb|680x680px|'''Fig. ISPa17.1.''' '''A)''' Map of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. The IS is represented as a yellow horizontal box with a scale showing the length in base pairs above. Open reading frames are shown as horizontal arrows below indicating the direction of expression: lilac indicated transposition-related ''orfs'' (an ''orf'' to the right closely related to the resolvase of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42''] and a second divergent ''orf'' with unknown function to the left). Between these are a toxin-antitoxin system (orange) identical to that of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'']. The terminal inverted repeats (IR) are indicated as horizontal grey arrows. The sequence of these is shown below. '''B)''' Similarities and difference between [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] and [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. The sequences of IRi and IRt are compared to those of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. The organization of the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ends together with the sub-repeats not carried by [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] is shown below. ]] | + | [[File:ISPa17-fig1.png|center|thumb|680x680px|'''Fig. ISPa17.1.''' '''A)''' Map of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. The IS is represented as a yellow horizontal box with a scale showing the length in base pairs above. Open reading frames are shown as horizontal arrows below indicating the direction of expression: lilac indicated transposition-related ''orfs'' (an ''orf'' to the right closely related to the resolvase of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42''] and a second divergent ''orf'' with unknown function to the left). Between these are a [[wikipedia:Toxin-antitoxin_system|toxin-antitoxin system]] (orange) identical to that of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'']. The terminal inverted repeats (IR) are indicated as horizontal grey arrows. The sequence of these is shown below. '''B)''' Similarities and difference between [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] and [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. The sequences of IRi and IRt are compared to those of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']. The organization of the [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] ends together with the sub-repeats not carried by [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] is shown below. ]] |
Interestingly, the IRs show almost complete identity with those of [[Transposons families/Tn402 family|Tn''402'' family transposons]] and their derivative integrons but does not include the multiple sub-repeats ([[:File:ISPa17-fig1.png|Fig. ISPa17.1]] '''B''') and [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] is often found associated with integron platforms (see: [[Transposons families/Tn3 family#Tn402 and integron platforms.|Tn''402'' and integron platforms]]). | Interestingly, the IRs show almost complete identity with those of [[Transposons families/Tn402 family|Tn''402'' family transposons]] and their derivative integrons but does not include the multiple sub-repeats ([[:File:ISPa17-fig1.png|Fig. ISPa17.1]] '''B''') and [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] is often found associated with integron platforms (see: [[Transposons families/Tn3 family#Tn402 and integron platforms.|Tn''402'' and integron platforms]]). | ||
====Association with integrons==== | ====Association with integrons==== | ||
| − | In one instance, a probable pathway for incorporation of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] into an integron structure has been documented <ref name=":1" | + | In one instance, a probable pathway for incorporation of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] into an integron structure has been documented <ref name=":1"><pubmed>23928025</pubmed></ref> ([[:File:ISPa17-fig2.png|Fig. ISPa17.2]] '''top'''). It was proposed that an ancestral target [[wikipedia:RK2_plasmid|IncP-1-ε plasmid]], pAKD25, acquired both [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] inserted within and inactivating the [[wikipedia:ParABS_system|''parA'' gene]] and an integron (Tn''402''-family top) insertion between ''[[wikipedia:ParABS_system|parA]]'' and [[wikipedia:Transfer_gene|''traC'' gene]]. Both insertions generated distinct 5 bp direct target repeats. This step is represented by pHH128 <ref name=":2"><pubmed>23831558</pubmed></ref> ([[:File:ISPa17-fig2.png|Fig. ISPa17.2]] '''middle'''). |
| − | It should be noted that pHH128 is part of a large family of [[wikipedia:RK2_plasmid|IncP-1 plasmids]], all with similar structures but with variations in the associated integron due to changes in the integron cassettes and insertion of various IS <ref name=":2" />. Pilato et al <ref name=":1" /> describe plasmid pAX22 (Fig. ISPa17.2) which carries an entire copy of the integron [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''], and it appears that, in this case, transposition into the target plasmid, pAX22, occurred simply using the IRt [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] end and the IR of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] as an alternative end to generate a 5 bp terminal DR (Fig. ISPa17.2 '''bottom'''). | + | It should be noted that pHH128 is part of a large family of [[wikipedia:RK2_plasmid|IncP-1-ε plasmids]], all with similar structures but with variations in the associated integron due to changes in the integron cassettes and insertion of various IS <ref name=":2" />. Pilato et al <ref name=":1" /> describe plasmid pAX22 ([[:File:ISPa17-fig2.png|Fig. ISPa17.2]]) which carries an entire copy of the integron [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''], and it appears that, in this case, transposition into the target plasmid, pAX22, occurred simply using the IRt [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] end and the IR of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] as an alternative end to generate a 5 bp terminal DR ([[:File:ISPa17-fig2.png|Fig. ISPa17.2]] '''bottom'''). |
This left the original 5bp “DR” at IR<sub>ISPa17</sub> and at the integron IRi. The [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] does not carry all the Tn''402''-associated ''tni'' genes necessary for autonomous transposition (see: [[Transposons families/Tn402 family#Tn402 Transposition-Related Genes|Tn''402'' Transposion-Related genes]]), although it has been established that transposition of such defective integron family TE can be accomplished by trans-complementation from an active [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-derivative in the same cell (see: [[Transposons families/Tn402 family|Tn''402'' family]]). Alternatively, transposition may be driven by the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] transposition machinery. It should be noted that [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] derivatives include sub-terminal repeats at both IRi and IRt, whereas the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] ends do not exhibit multiple sub-terminal repeats. It seems probable, then, that the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']-integron structure uses [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] transposition machinery. | This left the original 5bp “DR” at IR<sub>ISPa17</sub> and at the integron IRi. The [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] does not carry all the Tn''402''-associated ''tni'' genes necessary for autonomous transposition (see: [[Transposons families/Tn402 family#Tn402 Transposition-Related Genes|Tn''402'' Transposion-Related genes]]), although it has been established that transposition of such defective integron family TE can be accomplished by trans-complementation from an active [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']-derivative in the same cell (see: [[Transposons families/Tn402 family|Tn''402'' family]]). Alternatively, transposition may be driven by the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] transposition machinery. It should be noted that [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] derivatives include sub-terminal repeats at both IRi and IRt, whereas the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] ends do not exhibit multiple sub-terminal repeats. It seems probable, then, that the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']-integron structure uses [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] transposition machinery. | ||
| − | [[File:ISPa17-fig2.png|center|thumb|720x720px|'''Fig. ISPa17.2.''' '''Model of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']''' '''Assimilation into a Transposon Structure.''' Transposable elements are shown as horizontal yellow boxes and orfs as horizontal arrows. The ''par'' locus is shown in blue and ''traC'' in green. The Plasmid Genbank ID numbers are shown to the left. '''Top.''' Proposed target region of plasmid pAKD25. The target sequences for [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] (left) and the [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] integron (right) are indicated. Middle. Insertion of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and the integron. Plasmid pHH128 ''parA'' and ''traC'' region containing the insertions. The 5bp direct flanking target duplications are shown. Bottom. Resulting transposition of the integron and assimilated [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] copy. New flanking DNA in the plasmid pAX22 target is shown in red. The new flanking 5bp repeats are also shown. Note the retention of both internal [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] 5bp target sequences. Pilato et al.,<ref name=":1" />.]] | + | [[File:ISPa17-fig2.png|center|thumb|720x720px|'''Fig. ISPa17.2.''' '''Model of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']''' '''Assimilation into a Transposon Structure.''' Transposable elements are shown as horizontal yellow boxes and orfs as horizontal arrows. The ''par'' locus is shown in blue and ''traC'' in green. The Plasmid Genbank ID numbers are shown to the left. '''Top.''' Proposed target region of plasmid pAKD25. The target sequences for [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] (left) and the [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] integron (right) are indicated. Middle. Insertion of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and the integron. Plasmid pHH128 ''parA'' and ''traC'' region containing the insertions. The 5bp direct flanking target duplications are shown. Bottom. Resulting transposition of the integron and assimilated [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] copy. New flanking DNA in the plasmid pAX22 target is shown in red. The new flanking 5bp repeats are also shown. Note the retention of both internal [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] 5bp target sequences. Redraw and modified from Pilato et al.,<ref name=":1" />.]] |
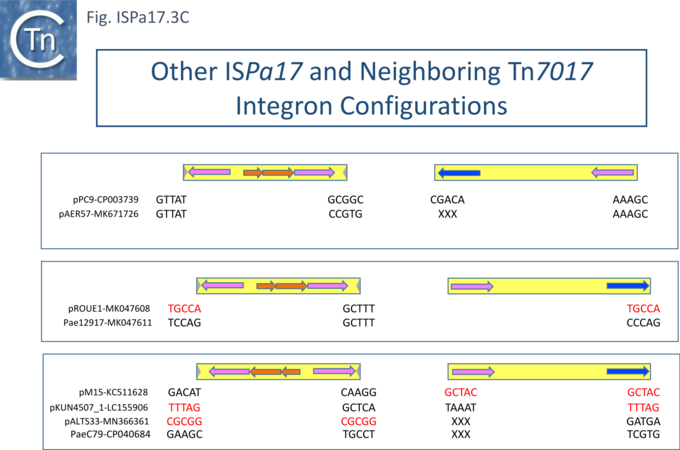
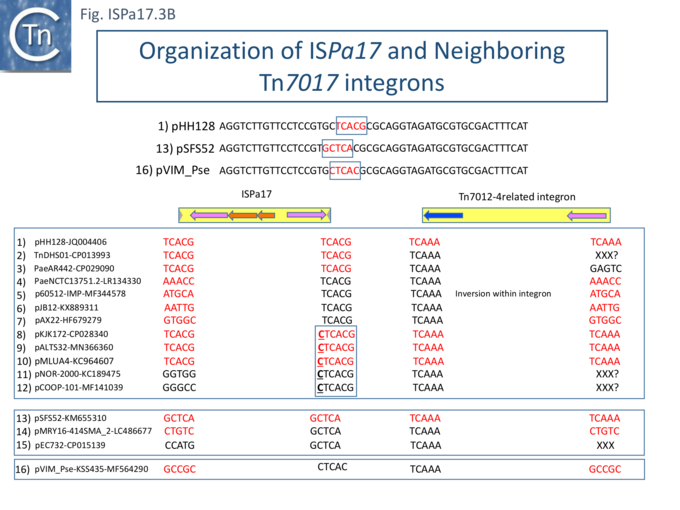
| − | Understanding [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] involvement in, and interaction with, integron recombination is not really understood in detail. Where [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] apparently occurs unlinked to an integron structure, it is often, but not always, flanked by 5bp direct repeats (Fig. ISPa17.3A). When it is linked to an integron, the majority appear to have integrated into the [[wikipedia:ParABS_system|''parA'' gene]] next to a particular integron insertion located between ''[[wikipedia:ParABS_system|parA]]'' and ''[[wikipedia:ParABS_system|traC]]'' of the host plasmid. The integron is flanked on both sides by a TCAAA repeat (line 1 and perhaps 2 since the integron IRt is not apparent Fig. ISPa17.3B). | + | Understanding [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] involvement in, and interaction with, integron recombination is not really understood in detail. Where [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] apparently occurs unlinked to an integron structure, it is often, but not always, flanked by 5bp direct repeats ([[:File:ISPa17-fig3A.png|Fig. ISPa17.3A]]). When it is linked to an integron, the majority appear to have integrated into the [[wikipedia:ParABS_system|''parA'' gene]] next to a particular integron insertion located between ''[[wikipedia:ParABS_system|parA]]'' and ''[[wikipedia:ParABS_system|traC]]'' of the host plasmid. The integron is flanked on both sides by a TCAAA repeat (line 1 and perhaps 2 since the integron IRt is not apparent ([[:File:ISPa17-fig3B.png|Fig. ISPa17.3B]]). |
| − | Here, the accompanying [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] copy is also flanked generally by a 5bp repeat, TCACG. It should be noted that, from one example to another, the integron may have undergone rearrangement, acquisition or loss of integron cassettes, insertions of other TE and TE-associated resulting in the disappearance of a target repeat (lines 2, 11 and 12). There are several examples (lines 5,6,7 and 8) which have clearly undergone transposition using the “''outside''” end of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and the integron IRt since the structure acquires different flanking 5bp but maintains the internal [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and IRi 5bp. (TCACG and TCAAA respectively). . | + | Here, the accompanying [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] copy is also flanked generally by a 5bp repeat, TCACG. It should be noted that, from one example to another, the integron may have undergone rearrangement, acquisition or loss of integron cassettes, insertions of other TE and TE-associated resulting in the disappearance of a target repeat (lines 2, 11 and 12). There are several examples (lines 5,6,7 and 8) which have clearly undergone transposition using the “''outside''” end of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and the integron IRt since the structure acquires different flanking 5bp but maintains the internal [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and IRi 5bp. (TCACG and TCAAA respectively). . |
The examples in lines 8-12 have acquired an additional C abutting the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] IR, but all retain the original 5bp repeat. The examples in lines 8, 9, and 10 therefore retain both sets of 5bp repeats. Those in line 11 and 12 may have undergone transposition of the entire [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']-Integron structure since the outside 5bp [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] has changed, but this could not be confirmed since the IRt integron ends could not be identified. | The examples in lines 8-12 have acquired an additional C abutting the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] IR, but all retain the original 5bp repeat. The examples in lines 8, 9, and 10 therefore retain both sets of 5bp repeats. Those in line 11 and 12 may have undergone transposition of the entire [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17'']-Integron structure since the outside 5bp [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] has changed, but this could not be confirmed since the IRt integron ends could not be identified. | ||
Clearly, all these derivatives have resulted from a single [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and integron insertion event. However, additional examples revealed two additional insertions into the [[wikipedia:ParABS_system|''parA'' gene]]: into GCTCA (line 13) and CTCAC (line 16). As for the example in line 1, that in line 13 also appears to have undergone transposition using the external ends of the IS-integron structure (line 14 and probably line 15). Note that the third integration site is implied from the two internal ends, and the element appears to have undergone transposition. | Clearly, all these derivatives have resulted from a single [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and integron insertion event. However, additional examples revealed two additional insertions into the [[wikipedia:ParABS_system|''parA'' gene]]: into GCTCA (line 13) and CTCAC (line 16). As for the example in line 1, that in line 13 also appears to have undergone transposition using the external ends of the IS-integron structure (line 14 and probably line 15). Note that the third integration site is implied from the two internal ends, and the element appears to have undergone transposition. | ||
| + | [[File:ISPa17-fig3A.png|center|thumb|640x640px|'''Fig. ISPa17.3A. Isolated IS''Pa17''.''' The organization of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] is indicated at the top showing the IS and associated orfs. The flanking 5bp are shown below: red indicated 5bp repeats. The different IS examples are shown with their source and genebank ID to the left. ]] | ||
| + | It should be noted that all three insertion targets overlap ([[:File:ISPa17-fig3B.png|Fig. ISPa17.3B]] '''top'''). This implies that the integron was inserted in an initial step, while the IS arrived subsequently. It is perhaps worthwhile to note that [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] copies are present in several ''[[wikipedia:Xanthomonas|Xanthomonas]]'' strains but do not appear to be associated with integrons and none were observed to be flanked by direct target repeats (Chandler unpublished observation). Further investigations are necessary to clarify this. | ||
| + | [[File:ISPa17-fig3B.png|center|thumb|680x680px|'''Fig. ISPa17.3B. Isolated IS''Pa17''. Top. Three insertion points for IS''Pa17'' within the ''parA'' gene.''' The 5bp target site which is duplicated on insertion is shown in red. '''Bottom. The organization of IS''Pa17'' and neighboring Tn''7017'' integrons.''' A cartoon of both is shown at the top. The flanking 5bp are shown below: red indicates 5bp repeats. The different examples are shown with their source and genebank ID to the left. ]] | ||
| + | [[File:ISPa17-fig3C.png|center|thumb|680x680px|'''Fig. ISPa17.3C. Isolated IS''Pa17''. Other IS''Pa17'' and Neighboring Tn''7017'' Integron Configurations.''' A cartoon of both is shown at the top. The flanking 5bp are shown below: red indicates 5bp repeats. The different examples are shown with their source and genebank ID to the left. ]] | ||
| + | ====Transposition mechanism==== | ||
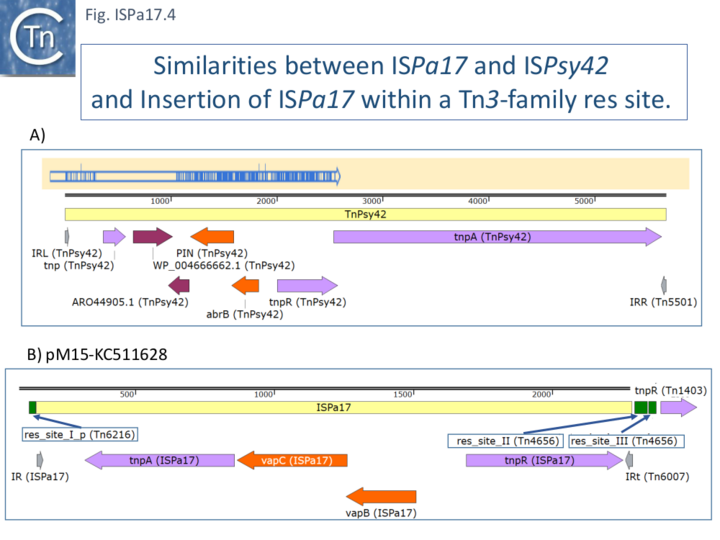
| + | Little is known about the [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] transposition mechanism. In addition to the [[wikipedia:Toxin-antitoxin_system|toxin/antitoxin]] genes, there are two divergent open reading frames. One shows remarkable identity to the resolvase of the [[Transposons families/Tn3 family|Tn''3'' family]] [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42''] (Identities: 180/187, 96%; Positive residues: 185/187, 99%; Gaps: 0/187, 0%) suggesting that transposition may involve a resolution step. | ||
| − | + | In addition, the [[wikipedia:Toxin-antitoxin_system|toxin/antitoxin]] genes show significant identity to those of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'']. The fourth ''orf'' shows some similarity to the N-terminal segment of the [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'' transposase] although there is no obvious similarity between the ends of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and those of [[Transposons families/Tn3 family|Tn''3'' family transposons]] ([[:File:ISPa17-fig3A.png|Fig. ISPa17.4A]]). | |
| + | It is well known that [https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402''] transposons show a strong propensity to integrate into [[Transposons families/Tn3 family|Tn''3'']] (and other) family ''res'' sites ([https://tncentral.ncc.unesp.br/report/te/Tn402-U67194.4 Tn''402'']). It seems possible that integration of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] involves targeting of ''res'' sites neighboring [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] derivatives. Moreover, there are several examples of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] insertion into ''res'' sites independently of [https://tncentral.ncc.unesp.br/report/te/Tn7017-NC_022242.1 Tn''7017''] ([[:File:ISPa17-fig3B.png|Fig. ISPa17.4B]]). | ||
| − | |||
| − | |||
| − | |||
<br /> | <br /> | ||
| − | + | [[File:ISPa17-fig4.png|center|thumb|720x720px|'''Fig. ISPa17.4. Top Similarities between [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42''].''' A cartoon of both is shown at the top. Transposons, yellow horizontal boxes, ''orfs'' horizontal arrows. Blue striated arrow indicates identities between [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 IS''Pa17''] and [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=ISPsy42 IS''Psy42'']. '''Bottom. Insertion of [https://tncentral.ncc.unesp.br/report/te/ISPa17-MF344578.1 ''ISPa17''] within a [[Transposons families/Tn3 family#Recombination at res.|Tn''3''-family ''res'' site]] (green boxes).''']] | |
| − | + | <br /> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Bibliography== | ==Bibliography== | ||
Latest revision as of 19:18, 23 April 2025
General
In contrast to other IS families, little attention has been paid to and element called ISPa17. It was first identified as an autonomous unit in the sequence of the IncP-6 plasmid Rms149 [1] along with a number of other potential TE and was shown as an element including 3 orfs. Haines et al note that it had been observed previously in the Pseudomonas aeruginosa plasmid pKLC102 as three orfs located downstream from an integron Int1 gene but not identified as an autonomous IS presumably because it was not flanked by a direct target repeat and did not exhibit a recognizable transposase [2].
The Rms149 copy was flanked by a 5 bp direct repeat (CGCGG) and was bordered by 25bp terminal inverted repeats (Fig. ISPa17.1A)[1]. It was located upstream from a Tn3-family transposase. Further analysis (Chandler unpublished observation) revealed 4 orfs, two of which constitute a toxin-antitoxin system related to vapBC gene which is also present in the Tn3 family transposon ISPsy42. A third orf was tentatively identified as a recombinase while a function for the fourth could not be identified.
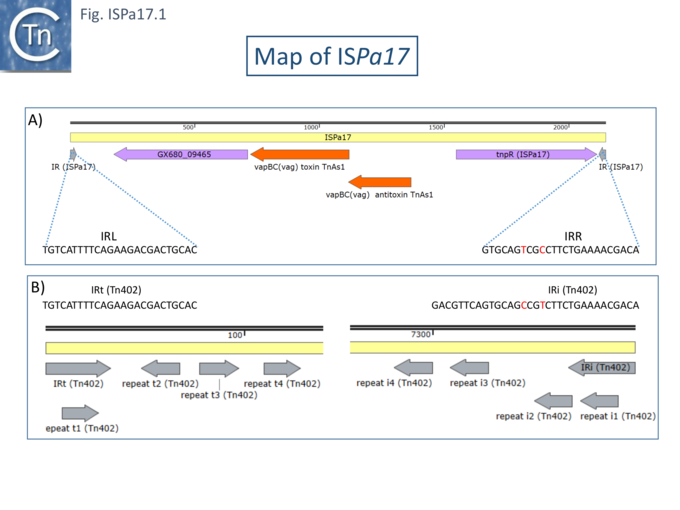
Interestingly, the IRs show almost complete identity with those of Tn402 family transposons and their derivative integrons but does not include the multiple sub-repeats (Fig. ISPa17.1 B) and ISPa17 is often found associated with integron platforms (see: Tn402 and integron platforms).
Association with integrons
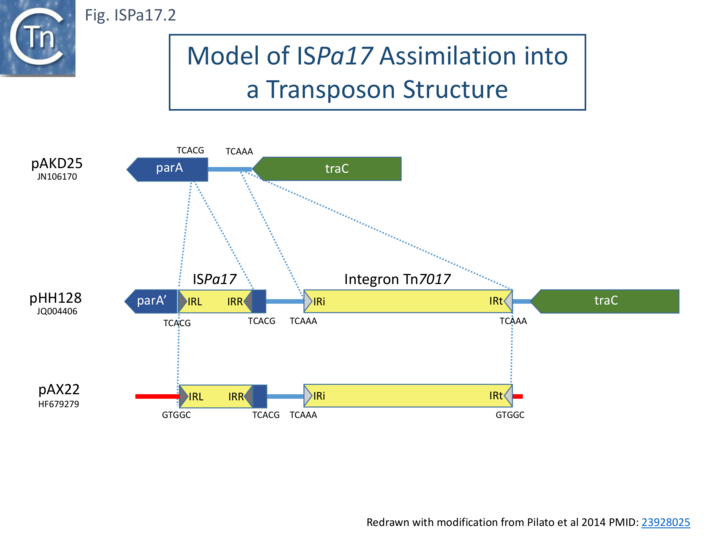
In one instance, a probable pathway for incorporation of ISPa17 into an integron structure has been documented [3] (Fig. ISPa17.2 top). It was proposed that an ancestral target IncP-1-ε plasmid, pAKD25, acquired both ISPa17 inserted within and inactivating the parA gene and an integron (Tn402-family top) insertion between parA and traC gene. Both insertions generated distinct 5 bp direct target repeats. This step is represented by pHH128 [4] (Fig. ISPa17.2 middle).
It should be noted that pHH128 is part of a large family of IncP-1-ε plasmids, all with similar structures but with variations in the associated integron due to changes in the integron cassettes and insertion of various IS [4]. Pilato et al [3] describe plasmid pAX22 (Fig. ISPa17.2) which carries an entire copy of the integron Tn7017, and it appears that, in this case, transposition into the target plasmid, pAX22, occurred simply using the IRt Tn7017 end and the IR of ISPa17 as an alternative end to generate a 5 bp terminal DR (Fig. ISPa17.2 bottom).
This left the original 5bp “DR” at IRISPa17 and at the integron IRi. The Tn7017 does not carry all the Tn402-associated tni genes necessary for autonomous transposition (see: Tn402 Transposion-Related genes), although it has been established that transposition of such defective integron family TE can be accomplished by trans-complementation from an active Tn402-derivative in the same cell (see: Tn402 family). Alternatively, transposition may be driven by the ISPa17 transposition machinery. It should be noted that Tn402 derivatives include sub-terminal repeats at both IRi and IRt, whereas the ISPa17 ends do not exhibit multiple sub-terminal repeats. It seems probable, then, that the ISPa17-integron structure uses ISPa17 transposition machinery.
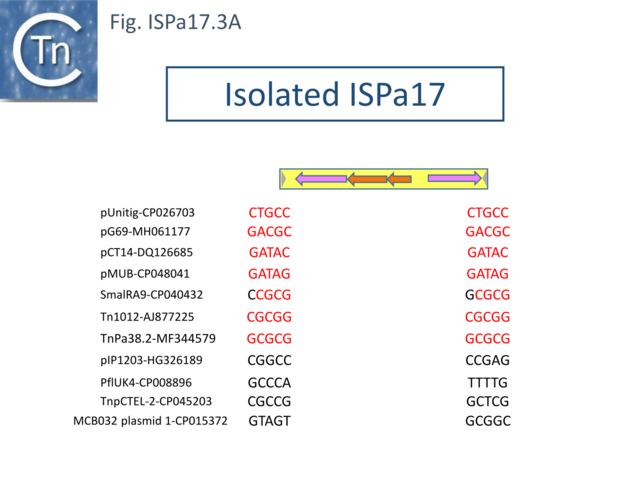
Understanding ISPa17 involvement in, and interaction with, integron recombination is not really understood in detail. Where ISPa17 apparently occurs unlinked to an integron structure, it is often, but not always, flanked by 5bp direct repeats (Fig. ISPa17.3A). When it is linked to an integron, the majority appear to have integrated into the parA gene next to a particular integron insertion located between parA and traC of the host plasmid. The integron is flanked on both sides by a TCAAA repeat (line 1 and perhaps 2 since the integron IRt is not apparent (Fig. ISPa17.3B).
Here, the accompanying ISPa17 copy is also flanked generally by a 5bp repeat, TCACG. It should be noted that, from one example to another, the integron may have undergone rearrangement, acquisition or loss of integron cassettes, insertions of other TE and TE-associated resulting in the disappearance of a target repeat (lines 2, 11 and 12). There are several examples (lines 5,6,7 and 8) which have clearly undergone transposition using the “outside” end of ISPa17 and the integron IRt since the structure acquires different flanking 5bp but maintains the internal ISPa17 and IRi 5bp. (TCACG and TCAAA respectively). .
The examples in lines 8-12 have acquired an additional C abutting the ISPa17 IR, but all retain the original 5bp repeat. The examples in lines 8, 9, and 10 therefore retain both sets of 5bp repeats. Those in line 11 and 12 may have undergone transposition of the entire ISPa17-Integron structure since the outside 5bp ISPa17 has changed, but this could not be confirmed since the IRt integron ends could not be identified.
Clearly, all these derivatives have resulted from a single ISPa17 and integron insertion event. However, additional examples revealed two additional insertions into the parA gene: into GCTCA (line 13) and CTCAC (line 16). As for the example in line 1, that in line 13 also appears to have undergone transposition using the external ends of the IS-integron structure (line 14 and probably line 15). Note that the third integration site is implied from the two internal ends, and the element appears to have undergone transposition.
It should be noted that all three insertion targets overlap (Fig. ISPa17.3B top). This implies that the integron was inserted in an initial step, while the IS arrived subsequently. It is perhaps worthwhile to note that ISPa17 copies are present in several Xanthomonas strains but do not appear to be associated with integrons and none were observed to be flanked by direct target repeats (Chandler unpublished observation). Further investigations are necessary to clarify this.
Transposition mechanism
Little is known about the ISPa17 transposition mechanism. In addition to the toxin/antitoxin genes, there are two divergent open reading frames. One shows remarkable identity to the resolvase of the Tn3 family ISPsy42 (Identities: 180/187, 96%; Positive residues: 185/187, 99%; Gaps: 0/187, 0%) suggesting that transposition may involve a resolution step.
In addition, the toxin/antitoxin genes show significant identity to those of ISPsy42. The fourth orf shows some similarity to the N-terminal segment of the ISPsy42 transposase although there is no obvious similarity between the ends of ISPa17 and those of Tn3 family transposons (Fig. ISPa17.4A).
It is well known that Tn402 transposons show a strong propensity to integrate into Tn3 (and other) family res sites (Tn402). It seems possible that integration of ISPa17 involves targeting of res sites neighboring Tn7017 derivatives. Moreover, there are several examples of ISPa17 insertion into res sites independently of Tn7017 (Fig. ISPa17.4B).
Bibliography
- ↑ 1.0 1.1 Haines AS, Jones K, Cheung M, Thomas CM . The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. - J Bacteriol: 2005 Jul, 187(14);4728-38 [PubMed:15995187] [DOI]
- ↑ Klockgether J, Reva O, Larbig K, Tümmler B . Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. - J Bacteriol: 2004 Jan, 186(2);518-34 [PubMed:14702321] [DOI]
- ↑ 3.0 3.1 3.2 Di Pilato V, Pollini S, Rossolini GM . Characterization of plasmid pAX22, encoding VIM-1 metallo-β-lactamase, reveals a new putative mechanism of In70 integron mobilization. - J Antimicrob Chemother: 2014 Jan, 69(1);67-71 [PubMed:23928025] [DOI]
- ↑ 4.0 4.1 Oliveira CS, Moura A, Henriques I, Brown CJ, Rogers LM, Top EM, Correia A . Comparative genomics of IncP-1ε plasmids from water environments reveals diverse and unique accessory genetic elements. - Plasmid: 2013 Nov, 70(3);412-9 [PubMed:23831558] [DOI]