Difference between revisions of "IS Families/IS481 family"
| Line 1: | Line 1: | ||
| + | ==== '''Historical''' ==== | ||
| + | IS''481'' was first identified in the late 1980s <ref><nowiki><pubmed>3426877</pubmed></nowiki></ref><ref name=":0"><nowiki><pubmed>2888834</pubmed></nowiki></ref> as a repeated sequence in the agent of whooping cough, ''[[wikipedia:Bordetella_pertussis|Bordetella pertussis]]''. It was estimated, from [[wikipedia:Southern_blot|Southern blots]] of restriction digested ''[[wikipedia:Bordetella_pertussis|B. pertusis]]'' genomic DNA, that [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS481 IS''481''] was present in multiple copies of about 20, with some variation between strains. They were not observed as repeated sequences in the single strain of ''[[wikipedia:Bordetella_parapertussis|B. parapertussis]]'' or in ''[[wikipedia:Bordetella_bronchiseptica|B. bronchiseptica]]'' which were analyzed, although homology was observed as a single band in ''[[wikipedia:Bordetella_parapertussis|B. parapertussis]]'' strain BPAH1 genomic digests and two bands in digests of [[wikipedia:Bordetella_bronchiseptica|''B. bronchiseptica'']] strain BBRH1. | ||
| + | |||
| + | The DNA sequence <ref name=":0" /><ref name=":1"><nowiki><pubmed>2908119</pubmed></nowiki></ref><ref><nowiki><pubmed>2552259</pubmed></nowiki></ref> later identified [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS481 IS''481''] as a 1052bp element with 28bp terminal inverted repeats and confirmed that it was not present in multiple copies in either the''[[wikipedia:Bordetella_parapertussis|B. parapertussis]]'' or [[wikipedia:Bordetella_bronchiseptica|''B. bronchiseptica'']] genomes. This was later reassessed to be 1046bp in length (Fig. IS481.1; <ref name=":2"><nowiki><pubmed>2229381</pubmed></nowiki></ref> and to massively hybridize with pertussis genomic DNA of 100 [[wikipedia:Bordetella_pertussis|''B. pertusis'']] clinical isolates. There were a number of small sequence variations observed in the results from different authors. | ||
| + | |||
| + | It was also observed to transpose. When the [https://www.uniprot.org/uniprotkb/P0A4H4/entry ''bvgA'' gene], a global virulence regulator, was present as both a plasmid and chromosomal copy, host growth was inhibited when the plasmid-borne gene carries a small deletion <ref><nowiki><pubmed>9573202</pubmed></nowiki></ref>. Those derivatives in which growth inhibition was relieved had acquired [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS481 IS''481''] insertions into the plasmid-borne gene or [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS481 IS''481'']/[https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS1002 IS''1002''] insertions into the chromosomal gene and to generate 6bp target DR <ref><nowiki><pubmed>9733704</pubmed></nowiki></ref>. | ||
| + | <br /> | ||
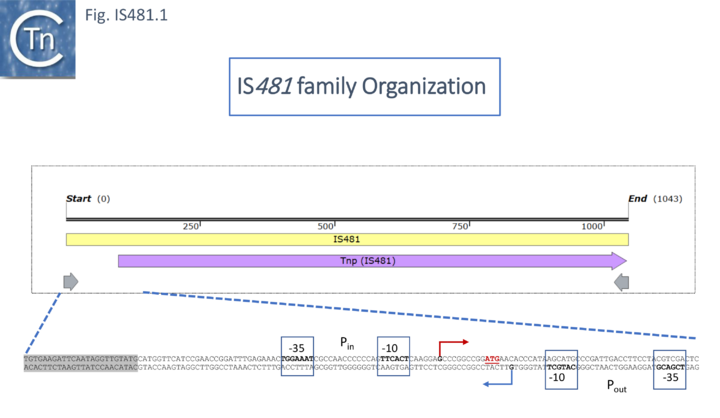
| + | [[File:IS481-fig1.png|center|thumb|720x720px|'''Fig. IS481.1. Organization of IS''481''. Top: A map of [https://tncentral.ncc.unesp.br/ISfinder/scripts/ficheIS.php?name=IS481 IS''481''].''' The insertion sequence is shown as a yellow horizontal box with its transposase gene as a lilac horizontal arrow to indicate the direction of expression. The 28 base pair inverted repeated are shown as horizontal gray arrows. '''Bottom: Transcriptional organization at the left IS end.''' The DNA sequence including the left inverted repeat (gray shading) together with the transposase promoter (Pin) oriented to the right with its -35 and -10 components labelled, the transcription start shown by a red arrow and the transposase initiation codon, '''ATG''', shown in red. An outward directed promoter Pout, overlapping with Pin and oriented to the left with its -35 and -10 components labelled, the transcription start shown by a blue arrow is also included <ref name=":1" /><ref name=":2" /><ref><nowiki><pubmed>39781897</pubmed></nowiki></ref>. ]] | ||
| + | |||
| + | |||
| + | '''Relationship to other IS.''' | ||
| + | |||
| + | Initially, IS''481'' appeared to be an IS''3'' family derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in 5’ TGT 3’) are similar to those of IS''3'' family members. (Fig. IS481.X???). Although, at that time IS''481'' had not been demonstrated to transpose, their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggested that these represent a distinct transpositionally active family. Different members generate DR of between 4 and 15 bp. Moreover, certain members (e.g. IS''Sav7'') insert specifically into the tetranucleotide CTAG which becomes the flanking DR and provides the UAG termination codon for the Tpase. In contrast to the vast majority of IS''3'' family members, the IS''481'' Tpase is not produced by frameshifting. There is no evidence for a leucine zipper as in IS''3.'' This can be clearly seen from the alignment and remarkable predicted structural similarities in the C-terminal catalytic domain and small N-terminal domain (Fig. IS481.2). | ||
| + | |||
| + | |||
====General==== | ====General==== | ||
'''<big>I</big>'''nitially, IS''481'' appeared to be an [[IS Families/IS3 family|IS''3'' family]] derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in '''5’''' TGT '''3’''') are similar to those of [[IS Families/IS3 family|IS''3'' family members]]. Their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggests that these represent a distinct transpositionally active family. | '''<big>I</big>'''nitially, IS''481'' appeared to be an [[IS Families/IS3 family|IS''3'' family]] derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in '''5’''' TGT '''3’''') are similar to those of [[IS Families/IS3 family|IS''3'' family members]]. Their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggests that these represent a distinct transpositionally active family. | ||
Revision as of 21:07, 22 April 2025
Historical
IS481 was first identified in the late 1980s [1][2] as a repeated sequence in the agent of whooping cough, Bordetella pertussis. It was estimated, from Southern blots of restriction digested B. pertusis genomic DNA, that IS481 was present in multiple copies of about 20, with some variation between strains. They were not observed as repeated sequences in the single strain of B. parapertussis or in B. bronchiseptica which were analyzed, although homology was observed as a single band in B. parapertussis strain BPAH1 genomic digests and two bands in digests of B. bronchiseptica strain BBRH1.
The DNA sequence [2][3][4] later identified IS481 as a 1052bp element with 28bp terminal inverted repeats and confirmed that it was not present in multiple copies in either theB. parapertussis or B. bronchiseptica genomes. This was later reassessed to be 1046bp in length (Fig. IS481.1; [5] and to massively hybridize with pertussis genomic DNA of 100 B. pertusis clinical isolates. There were a number of small sequence variations observed in the results from different authors.
It was also observed to transpose. When the bvgA gene, a global virulence regulator, was present as both a plasmid and chromosomal copy, host growth was inhibited when the plasmid-borne gene carries a small deletion [6]. Those derivatives in which growth inhibition was relieved had acquired IS481 insertions into the plasmid-borne gene or IS481/IS1002 insertions into the chromosomal gene and to generate 6bp target DR [7].
Relationship to other IS.
Initially, IS481 appeared to be an IS3 family derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in 5’ TGT 3’) are similar to those of IS3 family members. (Fig. IS481.X???). Although, at that time IS481 had not been demonstrated to transpose, their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggested that these represent a distinct transpositionally active family. Different members generate DR of between 4 and 15 bp. Moreover, certain members (e.g. ISSav7) insert specifically into the tetranucleotide CTAG which becomes the flanking DR and provides the UAG termination codon for the Tpase. In contrast to the vast majority of IS3 family members, the IS481 Tpase is not produced by frameshifting. There is no evidence for a leucine zipper as in IS3. This can be clearly seen from the alignment and remarkable predicted structural similarities in the C-terminal catalytic domain and small N-terminal domain (Fig. IS481.2).
General
Initially, IS481 appeared to be an IS3 family derivative which had been truncated for the N-terminal end of the Tpase and includes a C-terminal extension. The DDE active site domain and the IR (ending in 5’ TGT 3’) are similar to those of IS3 family members. Their presence in high copy number in some species and the identification of at least 130 distinct but related IS from over 90 species strongly suggests that these represent a distinct transpositionally active family.
Different members generate DR of between 4 and 15 bp. Moreover, certain members (e.g. ISSav7) insert specifically into the tetranucleotide CTAG which becomes the flanking DR and provides the UAG termination codon for the Tpase. In contrast to the vast majority of IS3 family members, the IS481 Tpase is not produced by frameshifting. There is no evidence for a leucine zipper as in IS3.
Some members include passenger genes including antibiotic resistance (Chloramphenicol - CmR gene for IS5564 and ISCgl1), or potential transcriptional regulators (ISKrh1, ISPfr21, ISSav7). IS481 itself has played a fundamental role in the evolution of the genomes of the Bordetellae where, in B. pertusis it has undergone extensive amplification to several hundred copies with accompanying genome decay[9][10] (Fig.IS481.1),
These IS are distantly related to the eukaryotic Banshee transposon which at present is restricted to the anaerobic flagellated protozoan Trichomonas vaginalis (Pritham per. comm.)[11]. They share the highly conserved Pfam integrase core domain identified initially in the IS3 family and retroviruses[12][13]. They also show a conserved 5’ TG 3’ tip to the IR which is typical of this and other types of mobile elements. It would be interesting to determine whether Banshee transposes using a dsDNA circular intermediate as do IS3 family members.
IS1202 group (ISNCY)
A small group including IS1202[14], which had been included in the ISNCY (Not Classified Yet) group appears distantly related to IS481. Members are between 1400 and 1700 bp (except for ISKpn21 which includes a passenger gene annotated as "hypothetical protein”) with a Tpase orf of between 400 and 500 aa in a single reading frame. Their IR begin with TGT as do those of the IS3 and IS481 families. They generate DR of between 5 and, unusually, 27 bp.
They appear to have similarities at the level of their Tpases particularly in their DDE domains (e.g. IS1202 is 39% aa similar to ISPfr5 of the IS481 family). Furthermore, they include glutamine (Q) seven residues C-terminal to the conserved E instead of the characteristic K/R. Identification of additional IS will be necessary to clearly define this group.
Bibliography